CEST MR fingerprinting (CEST-MRF) for brain tumor quantification using EPI readout and deep learning reconstruction
- PMID: 36128888
- PMCID: PMC9617776
- DOI: 10.1002/mrm.29448
CEST MR fingerprinting (CEST-MRF) for brain tumor quantification using EPI readout and deep learning reconstruction
Abstract
Purpose: To develop a clinical CEST MR fingerprinting (CEST-MRF) method for brain tumor quantification using EPI acquisition and deep learning reconstruction.
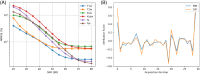
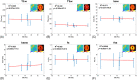
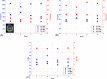
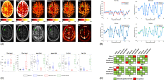
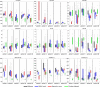
Methods: A CEST-MRF pulse sequence originally designed for animal imaging was modified to conform to hardware limits on clinical scanners while keeping scan time under 2 min. Quantitative MRF reconstruction was performed using a deep reconstruction network (DRONE) to yield the water relaxation and chemical exchange parameters. The feasibility of the six parameter DRONE reconstruction was tested in simulations using a digital brain phantom. A healthy subject was scanned with the CEST-MRF sequence, conventional MRF and CEST sequences for comparison. Reproducibility was assessed via test-retest experiments and the concordance correlation coefficient calculated for white matter and gray matter. The clinical utility of CEST-MRF was demonstrated on four patients with brain metastases in comparison to standard clinical imaging sequences. Tumors were segmented into edema, solid core, and necrotic core regions and the CEST-MRF values compared to the contra-lateral side.
Results: DRONE reconstruction of the digital phantom yielded a normalized RMS error of ≤7% for all parameters. The CEST-MRF parameters were in good agreement with those from conventional MRF and CEST sequences and previous studies. The mean concordance correlation coefficient for all six parameters was 0.98 ± 0.01 in white matter and 0.98 ± 0.02 in gray matter. The CEST-MRF values in nearly all tumor regions were significantly different (P = 0.05) from each other and the contra-lateral side.
Conclusion: Combination of EPI readout and deep learning reconstruction enabled fast, accurate and reproducible CEST-MRF in brain tumors.
Keywords: DRONE; chemical exchange rate; chemical exchange saturation transfer (CEST); deep learning; magnetic resonance fingerprinting (MRF); pH.
© 2022 The Authors. Magnetic Resonance in Medicine published by Wiley Periodicals LLC on behalf of International Society for Magnetic Resonance in Medicine.
Conflict of interest statement
O.C. and C.T.F. hold patents on the CEST‐MRF technology.
Figures



References
-
- Longo DL, Bartoli A, Consolino L, et al. In vivo imaging of tumor metabolism and acidosis by combining PET and MRI‐CEST pH imaging. Cancer Res. 2016;76:6463‐6470. - PubMed
-
- Granja S, Tavares‐Valente D, Queiros O, Baltazar F. Value of pH regulators in the diagnosis, prognosis and treatment of cancer. Semin Cancer Biol. 2017;43:17‐34. - PubMed
-
- Parks SK, Chiche J, Pouysségur J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat Rev Cancer. 2013;13:611‐623. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

