Determining the Likelihood of Disease Pathogenicity Among Incidentally Identified Genetic Variants in Rare Dilated Cardiomyopathy-Associated Genes
- PMID: 36129056
- PMCID: PMC9673717
- DOI: 10.1161/JAHA.122.025257
Determining the Likelihood of Disease Pathogenicity Among Incidentally Identified Genetic Variants in Rare Dilated Cardiomyopathy-Associated Genes
Abstract
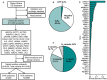
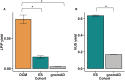
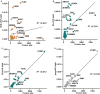
Background As utilization of clinical exome sequencing (ES) has expanded, criteria for evaluating the diagnostic weight of incidentally identified variants are critical to guide clinicians and researchers. This is particularly important in genes associated with dilated cardiomyopathy (DCM), which can cause heart failure and sudden death. We sought to compare the frequency and distribution of incidentally identified variants in DCM-associated genes between a clinical referral cohort with those in control and known case cohorts to determine the likelihood of pathogenicity among those undergoing genetic testing for non-DCM indications. Methods and Results A total of 39 rare, non-TTN DCM-associated genes were identified and evaluated from a clinical ES testing referral cohort (n=14 005, Baylor Genetic Laboratories) and compared with a DCM case cohort (n=9442) as well as a control cohort of population variants (n=141 456) derived from the gnomAD database. Variant frequencies in each cohort were compared. Signal-to-noise ratios were calculated comparing the DCM and ES cohort with the gnomAD cohort. The likely pathogenic/pathogenic variant yield in the DCM cohort (8.2%) was significantly higher than in the ES cohort (1.9%). Based on signal-to-noise and correlation analysis, incidental variants found in FLNC, RBM20, MYH6, DSP, ABCC9, JPH2, and NEXN had the greatest chance of being DCM-associated. Conclusions The distribution of pathogenic variants between the ES cohort and the DCM case cohort was gene specific, and variants found in the ES cohort were similar to variants found in the control cohort. Incidentally identified variants in specific genes are more associated with DCM than others.
Keywords: dilated cardiomyopathy; exome sequencing; genetics; incidental finding; secondary finding.
Figures



Similar articles
-
Reevaluating the Genetic Contribution of Monogenic Dilated Cardiomyopathy.Circulation. 2020 Feb 4;141(5):387-398. doi: 10.1161/CIRCULATIONAHA.119.037661. Epub 2020 Jan 27. Circulation. 2020. PMID: 31983221 Free PMC article.
-
Signal-to-Noise Analysis Can Inform the Likelihood That Incidentally Identified Variants in Sarcomeric Genes Are Associated with Pediatric Cardiomyopathy.J Pers Med. 2022 Apr 30;12(5):733. doi: 10.3390/jpm12050733. J Pers Med. 2022. PMID: 35629155 Free PMC article.
-
Incidentally identified genetic variants in arrhythmogenic right ventricular cardiomyopathy-associated genes among children undergoing exome sequencing reflect healthy population variation.Mol Genet Genomic Med. 2019 Jun;7(6):e593. doi: 10.1002/mgg3.593. Epub 2019 Apr 15. Mol Genet Genomic Med. 2019. PMID: 30985088 Free PMC article.
-
Biallelic NEXN variants and fetal onset dilated cardiomyopathy: two independent case reports and revision of literature.Ital J Pediatr. 2024 Aug 26;50(1):156. doi: 10.1186/s13052-024-01678-x. Ital J Pediatr. 2024. PMID: 39183344 Free PMC article. Review.
-
Arrhythmic Genotypes in Familial Dilated Cardiomyopathy: Implications for Genetic Testing and Clinical Management.Heart Lung Circ. 2019 Jan;28(1):31-38. doi: 10.1016/j.hlc.2018.09.010. Epub 2018 Oct 11. Heart Lung Circ. 2019. PMID: 30482687 Review.
Cited by
-
Rapid progression of right ventricular dysfunction: a case report.BMC Cardiovasc Disord. 2025 Mar 7;25(1):157. doi: 10.1186/s12872-025-04601-2. BMC Cardiovasc Disord. 2025. PMID: 40055607 Free PMC article.
-
Extending the Mutational Spectrum of SYNE1 Ataxia in Chinese Patients.Cerebellum. 2025 Aug 22;24(5):144. doi: 10.1007/s12311-025-01898-9. Cerebellum. 2025. PMID: 40841461 No abstract available.
-
Evaluating the functional and genomic analysis of pathogenic junctophilin-2 variants and their association with the pathogenesis of cardiomyopathy to understand their molecular impact on cardiac calcium homeostasis and disease phenotypes.Ann Pediatr Cardiol. 2024 Nov-Dec;17(6):401-407. doi: 10.4103/apc.apc_173_24. Epub 2025 Apr 24. Ann Pediatr Cardiol. 2024. PMID: 40352421 Free PMC article. Review.
-
Identification of BMP10 as a Novel Gene Contributing to Dilated Cardiomyopathy.Diagnostics (Basel). 2023 Jan 9;13(2):242. doi: 10.3390/diagnostics13020242. Diagnostics (Basel). 2023. PMID: 36673052 Free PMC article.
-
The clinical and genetic spectrum of pediatric hypertrophic cardiomyopathy manifesting before one year of age.Pediatr Res. 2025 Mar 18. doi: 10.1038/s41390-025-03989-z. Online ahead of print. Pediatr Res. 2025. PMID: 40102575
References
-
- Pinto YM, Elliott PM, Arbustini E, Adler Y, Anastasakis A, Böhm M, Duboc D, Gimeno J, de Groote P, Imazio M, et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non‐dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. 2016;37:1850–1858. doi: 10.1093/eurheartj/ehv727 - DOI - PubMed
-
- Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JB. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association scientific statement from the council on clinical cardiology, heart failure and transplantation committee; quality of care and outcomes research and functional genomics and translational biology interdisciplinary working groups; and council on epidemiology and prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

