Haplotype sequence collection of ABO blood group alleles by long-read sequencing reveals putative A1-diagnostic variants
- PMID: 36129841
- PMCID: PMC10025113
- DOI: 10.1182/bloodadvances.2022007133
Haplotype sequence collection of ABO blood group alleles by long-read sequencing reveals putative A1-diagnostic variants
Abstract
In the era of blood group genomics, reference collections of complete and fully resolved blood group gene alleles have gained high importance. For most blood groups, however, such collections are currently lacking, as resolving full-length gene sequences as haplotypes (ie, separated maternal/paternal origin) remains exceedingly difficult with both Sanger and short-read next-generation sequencing. Using the latest third-generation long-read sequencing, we generated a collection of fully resolved sequences for all 6 main ABO allele groups: ABO∗A1/A2/B/O.01.01/O.01.02/O.02. We selected 77 samples from an ABO genotype data set (n = 25 200) of serologically typed Swiss blood donors. The entire ABO gene was amplified in 2 overlapping long-range polymerase chain reactions (covering ∼23.6 kb) and sequenced by long-read Oxford Nanopore sequencing. For quality validation, 2 samples per ABO group were resequenced using Illumina and Pacific Biosciences technology. All 154 full-length ABO sequences were resolved as haplotypes. We observed novel, distinct sequence patterns for each ABO group. Most genetic diversity was found between, not within, ABO groups. Phylogenetic tree and haplotype network analyses highlighted distinct clades of each ABO group. Strikingly, our data uncovered 4 genetic variants putatively specific for ABO∗A1, for which direct diagnostic targets are currently lacking. We validated A1-diagnostic potential using whole-genome data (n = 4872) of a multiethnic cohort. Overall, our sequencing strategy proved powerful for producing high-quality ABO haplotypes and holds promise for generating similar collections for other blood groups. The publicly available collection of 154 haplotypes will serve as a valuable resource for molecular analyses of ABO, as well as studies about the function and evolutionary history of ABO.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: C.G. acts as a consultant for inno-train GmbH, Kronberg im Taunus, Germany. The remaining authors declare no competing financial interests.
Figures


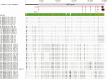
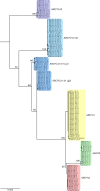
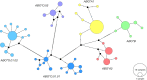
References
-
- Wheeler MM, Johnsen JM. The role of genomics in transfusion medicine. Curr Opin Hematol. 2018;25(6):509–515. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

