Higher-order epistasis and phenotypic prediction
- PMID: 36129941
- PMCID: PMC9522415
- DOI: 10.1073/pnas.2204233119
Higher-order epistasis and phenotypic prediction
Abstract
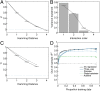
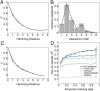
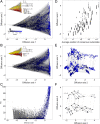
Contemporary high-throughput mutagenesis experiments are providing an increasingly detailed view of the complex patterns of genetic interaction that occur between multiple mutations within a single protein or regulatory element. By simultaneously measuring the effects of thousands of combinations of mutations, these experiments have revealed that the genotype-phenotype relationship typically reflects not only genetic interactions between pairs of sites but also higher-order interactions among larger numbers of sites. However, modeling and understanding these higher-order interactions remains challenging. Here we present a method for reconstructing sequence-to-function mappings from partially observed data that can accommodate all orders of genetic interaction. The main idea is to make predictions for unobserved genotypes that match the type and extent of epistasis found in the observed data. This information on the type and extent of epistasis can be extracted by considering how phenotypic correlations change as a function of mutational distance, which is equivalent to estimating the fraction of phenotypic variance due to each order of genetic interaction (additive, pairwise, three-way, etc.). Using these estimated variance components, we then define an empirical Bayes prior that in expectation matches the observed pattern of epistasis and reconstruct the genotype-phenotype mapping by conducting Gaussian process regression under this prior. To demonstrate the power of this approach, we present an application to the antibody-binding domain GB1 and also provide a detailed exploration of a dataset consisting of high-throughput measurements for the splicing efficiency of human pre-mRNA [Formula: see text] splice sites, for which we also validate our model predictions via additional low-throughput experiments.
Keywords: Gaussian processes; genetic interaction; genotype–phenotype map; protein G; splicing.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Kondrashov D. A., Kondrashov F. A., Topological features of rugged fitness landscapes in sequence space. Trends Genet. 31, 24–33 (2015). - PubMed
-
- Domingo J., Baeza-Centurion P., Lehner B., The causes and consequences of genetic interactions (epistasis). Annu. Rev. Genomics Hum. Genet. 20, 433–460 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

