d-StructMAn: Containerized structural annotation on the scale from genetic variants to whole proteomes
- PMID: 36130085
- PMCID: PMC9487898
- DOI: 10.1093/gigascience/giac086
d-StructMAn: Containerized structural annotation on the scale from genetic variants to whole proteomes
Abstract
Background: Structural annotation of genetic variants in the context of intermolecular interactions and protein stability can shed light onto mechanisms of disease-related phenotypes. Three-dimensional structures of related proteins in complexes with other proteins, nucleic acids, or ligands enrich such functional interpretation, since intermolecular interactions are well conserved in evolution.
Results: We present d-StructMAn, a novel computational method that enables structural annotation of local genetic variants, such as single-nucleotide variants and in-frame indels, and implements it in a highly efficient and user-friendly tool provided as a Docker container. Using d-StructMAn, we annotated several very large sets of human genetic variants, including all variants from ClinVar and all amino acid positions in the human proteome. We were able to provide annotation for more than 46% of positions in the human proteome representing over 60% proteins.
Conclusions: d-StructMAn is the first of its kind and a highly efficient tool for structural annotation of protein-coding genetic variation in the context of observed and potential intermolecular interactions. d-StructMAn is readily applicable to proteome-scale datasets and can be an instrumental building machine-learning tool for predicting genotype-to-phenotype relationships.
Keywords: Docker container; Single-nucleotide variants; genetic variation; indels; protein interactions; protein structure; structural annotation.
© The Author(s) 2022. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare no competing interests.
Figures



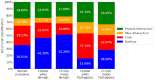

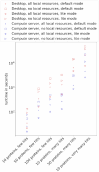
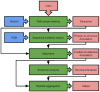
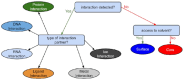

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

