HIV-1-Infected CD4+ T Cells Present MHC Class II-Restricted Epitope via Endogenous Processing
- PMID: 36130133
- PMCID: PMC9512365
- DOI: 10.4049/jimmunol.2200145
HIV-1-Infected CD4+ T Cells Present MHC Class II-Restricted Epitope via Endogenous Processing
Abstract
HIV-1-specific CD4+ T cells (TCD4+s) play a critical role in controlling HIV-1 infection. Canonically, TCD4+s are activated by peptides derived from extracellular ("exogenous") Ags displayed in complex with MHC class II (MHC II) molecules on the surfaces of "professional" APCs such as dendritic cells (DCs). In contrast, activated human TCD4+s, which express MHC II, are not typically considered for their APC potential because of their low endocytic capacity and the exogenous Ag systems historically used for assessment. Using primary TCD4+s and monocyte-derived DCs from healthy donors, we show that activated human TCD4+s are highly effective at MHC II-restricted presentation of an immunodominant HIV-1-derived epitope postinfection and subsequent noncanonical processing and presentation of endogenously produced Ag. Our results indicate that, in addition to marshalling HIV-1-specific immune responses during infection, TCD4+s also act as APCs, leading to the activation of HIV-1-specific TCD4+s.
Copyright © 2022 by The American Association of Immunologists, Inc.
Figures
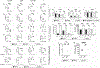



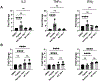
References
-
- UNAIDS. 2021. Global HIV & AIDS statistics — Fact sheet.
-
- Raynaud-Messina B, Bracq L, Dupont M, Souriant S, Usmani SM, Proag A, Pingris K, Soldan V, Thibault C, Capilla F, Al Saati T, Gennero I, Jurdic P, Jolicoeur P, Davignon JL, Mempel TR, Benichou S, Maridonneau-Parini I, and Verollet C. 2018. Bone degradation machinery of osteoclasts: An HIV-1 target that contributes to bone loss. Proc Natl Acad Sci U S A 115: E2556–E2565. - PMC - PubMed
-
- Moir S, Chun TW, and Fauci AS. 2011. Pathogenic mechanisms of HIV disease. Annu Rev Pathol 6: 223–248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

