Aryl Fluorosulfate Based Inhibitors That Covalently Target the SIRT5 Lysine Deacylase
- PMID: 36130196
- PMCID: PMC9828517
- DOI: 10.1002/anie.202204565
Aryl Fluorosulfate Based Inhibitors That Covalently Target the SIRT5 Lysine Deacylase
Abstract
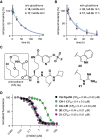
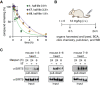
The sirtuin enzymes are a family of lysine deacylases that regulate gene transcription and metabolism. Sirtuin 5 (SIRT5) hydrolyzes malonyl, succinyl, and glutaryl ϵ-N-carboxyacyllysine posttranslational modifications and has recently emerged as a vulnerability in certain cancers. However, chemical probes to illuminate its potential as a pharmacological target have been lacking. Here we report the harnessing of aryl fluorosulfate-based electrophiles as an avenue to furnish covalent inhibitors that target SIRT5. Alkyne-tagged affinity-labeling agents recognize and capture overexpressed SIRT5 in cultured HEK293T cells and can label SIRT5 in the hearts of mice upon intravenous injection of the compound. This work demonstrates the utility of aryl fluorosulfate electrophiles for targeting of SIRT5 and suggests this as a means for the development of potential covalent drug candidates. It is our hope that these results will serve as inspiration for future studies investigating SIRT5 and general sirtuin biology in the mitochondria.
Keywords: Covalent Inhibition; Enzyme Inhibitors; SIRT5; Sirtuins; SuFEx.
© 2022 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

