Spanish HTT gene study reveals haplotype and allelic diversity with possible implications for germline expansion dynamics in Huntington disease
- PMID: 36130218
- PMCID: PMC9990985
- DOI: 10.1093/hmg/ddac224
Spanish HTT gene study reveals haplotype and allelic diversity with possible implications for germline expansion dynamics in Huntington disease
Abstract
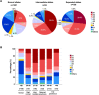
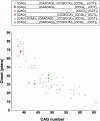
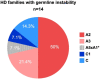
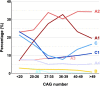
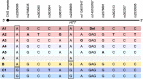
We aimed to determine the genetic diversity and molecular characteristics of the Huntington disease (HD) gene (HTT) in Spain. We performed an extended haplotype and exon one deep sequencing analysis of the HTT gene in a nationwide cohort of population-based controls (n = 520) and families with symptomatic individuals referred for HD genetic testing. This group included 331 HD cases and 140 carriers of intermediate alleles. Clinical and family history data were obtained when available. Spanish normal alleles are enriched in C haplotypes (40.1%), whereas A1 (39.8%) and A2 (31.6%) prevail among intermediate and expanded alleles, respectively. Alleles ≥ 50 CAG repeats are primarily associated with haplotypes A2 (38.9%) and C (32%), which are also present in 50% and 21.4%, respectively, of HD families with large intergenerational expansions. Non-canonical variants of exon one sequence are less frequent, but much more diverse, in alleles of ≥27 CAG repeats. The deletion of CAACAG, one of the six rare variants not observed among smaller normal alleles, is associated with haplotype C and appears to correlate with larger intergenerational expansions and early onset of symptoms. Spanish HD haplotypes are characterized by a high genetic diversity, potentially admixed with other non-Caucasian populations, with a higher representation of A2 and C haplotypes than most European populations. Differences in haplotype distributions across the CAG length range support differential germline expansion dynamics, with A2 and C showing the largest intergenerational expansions. This haplotype-dependent germline instability may be driven by specific cis-elements, such as the CAACAG deletion.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
-
- Rubinsztein, D.C., Leggo, J., Coles, R., Almqvist, E., Biancalana, V., Cassiman, J.-J., Chotai, K., Connarty, M., Craufurd, D., Curtis, A. et al. (1996) Phenotypic characterization of individuals with 30-40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36-39 repeats. Am. J. Hum. Genet., 59, 16–22. - PMC - PubMed
-
- Semaka, A., Collins, J.A. and Hayden, M.R. (2010) Unstable familial transmissions of huntington disease alleles with 27-35 CAG repeats (intermediate alleles). Am. J. Med. Genet. Part B Neuropsychiatr. Genet., 153, 314–320. - PubMed
-
- Rawlins, M.D., Wexler, N.S., Wexler, A.R., Tabrizi, S.J., Douglas, I., Evans, S.J.W. and Smeeth, L. (2016) The prevalence of Huntington’s disease. Neuroepidemiology, 46, 144–153. - PubMed
-
- Kay, C., Collins, J.A., Wright, G.E.B., Baine, F., Miedzybrodzka, Z., Aminkeng, F., Semaka, A.J., McDonald, C., Davidson, M., Madore, S.J. et al. (2018) The molecular epidemiology of Huntington disease is related to intermediate allele frequency and haplotype in the general population. Am. J. Med. Genet. Part B Neuropsychiatr. Genet., 177, 346–357. - PubMed
-
- Warby, S.C., Montpetit, A., Hayden, A.R., Carroll, J.B., Butland, S.L., Visscher, H., Collins, J.A., Semaka, A., Hudson, T.J. and Hayden, M.R. (2009) CAG Expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet., 84, 351–366. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

