Human anti-smallpox long-lived memory B cells are defined by dynamic interactions in the splenic niche and long-lasting germinal center imprinting
- PMID: 36130603
- PMCID: PMC7613742
- DOI: 10.1016/j.immuni.2022.08.019
Human anti-smallpox long-lived memory B cells are defined by dynamic interactions in the splenic niche and long-lasting germinal center imprinting
Abstract
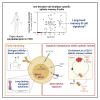
Memory B cells (MBCs) can persist for a lifetime, but the mechanisms that allow their long-term survival remain poorly understood. Here, we isolated and analyzed human splenic smallpox/vaccinia protein B5-specific MBCs in individuals who were vaccinated more than 40 years ago. Only a handful of clones persisted over such an extended period, and they displayed limited intra-clonal diversity with signs of extensive affinity-based selection. These long-lived MBCs appeared enriched in a CD21hiCD20hi IgG+ splenic B cell subset displaying a marginal-zone-like NOTCH/MYC-driven signature, but they did not harbor a unique longevity-associated transcriptional or metabolic profile. Finally, the telomeres of B5-specific, long-lived MBCs were longer than those in patient-paired naive B cells in all the samples analyzed. Overall, these results imply that separate mechanisms such as early telomere elongation, affinity selection during the contraction phase, and access to a specific niche contribute to ensuring the functional longevity of MBCs.
Keywords: affinity selection; long-lasting immune memory; memory B cells; smallpox; splenic microenvironment; telomeres; vaccinia.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.M. received research funds from GSK and personal fees from LFB and Amgen, and J.-C.W. received consulting fees from Institut Mérieux, all outside of the submitted work.
Figures







References
-
- Aldaz-Carroll L, Whitbeck JC, Ponce de Leon M, Lou H, Hirao L, Isaacs SN, Moss B, Eisenberg RJ, Cohen GH. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R. J Virol. 2005;79:6260–6271. doi: 10.1128/JVI.79.10.6260-6271.2005. - DOI - PMC - PubMed

