OCT4 interprets and enhances nucleosome flexibility
- PMID: 36130732
- PMCID: PMC9561370
- DOI: 10.1093/nar/gkac755
OCT4 interprets and enhances nucleosome flexibility
Abstract
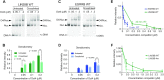
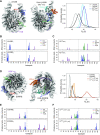
Pioneer transcription factors are proteins that induce cellular identity transitions by binding to inaccessible regions of DNA in nuclear chromatin. They contribute to chromatin opening and recruit other factors to regulatory DNA elements. The structural features and dynamics modulating their interaction with nucleosomes are still unresolved. From a combination of experiments and molecular simulations, we reveal here how the pioneer factor and master regulator of pluripotency, Oct4, interprets and enhances nucleosome structural flexibility. The magnitude of Oct4's impact on nucleosome dynamics depends on the binding site position and the mobility of the unstructured tails of nucleosomal histone proteins. Oct4 uses both its DNA binding domains to propagate and stabilize open nucleosome conformations, one for specific sequence recognition and the other for nonspecific interactions with nearby regions of DNA. Our findings provide a structural basis for the versatility of transcription factors in engaging with nucleosomes and have implications for understanding how pioneer factors induce chromatin dynamics.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389:251–260. - PubMed
-
- Cramer P. Organization and regulation of gene transcription. Nature. 2019; 573:45–54. - PubMed
-
- Cirillo L.A., Lin F.R., Cuesta I., Friedman D., Jarnik M., Zaret K.S.. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002; 9:279–289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

