Identification of rs2736099 as a novel cis-regulatory variation for TERT and implications for tumorigenesis and cell proliferation
- PMID: 36131156
- PMCID: PMC11797200
- DOI: 10.1007/s00432-022-04372-9
Identification of rs2736099 as a novel cis-regulatory variation for TERT and implications for tumorigenesis and cell proliferation
Abstract
Purpose: Lung cancer is a malignant tumor with obvious genetic predisposition. Association studies have proposed that rs2853677, a SNP localizing at intron region of TERT (telomerase reverse transcriptase), is significantly associated with TERT expression, telomere length and eventually lung cancer risk. However, functional genomics work indicates that rs2853677 is not with the ability to alter gene expression. All these facts make us hypothesize that some other genetic variation(s) are in linkage disequilibrium (LD) with rs2853677 and influence TERT expression.
Methods: LD pattern in rs2853677 nearby region was analyzed based on 1000 genomes data for three representative populations in the world and functional genomics research was performed for this locus.
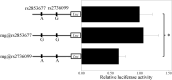
Results: Only one SNP, rs2736099, is in strong LD with rs2853677 in East Asian. Dual-luciferase reporter assay verifies that rs2736099 can regulate gene expression and should be the causal SNP for this disease. Through chromosome conformation capture assay, it is disclosed that the enhancer surrounding rs2736099 can interact with TERT promoter. Through chromatin immunoprecipitation, the transcription factor SP1 (Sp1 transcription factor) is recognized for the chromatin segment spanning rs2736099.
Conclusions: Our results provide the missing piece between genetic variation at this locus and lung cancer risk, which is also applied to tumorigenesis in other tissues and cell proliferation.
Keywords: Expression regulation; Lung cancer; TERT; rs2736099; rs2853677.
© 2022. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures



References
-
- Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima MA, Lambourne JJ, Sivapalaratnam S, Downes K, Kundu K, Bomba L, Berentsen K, Bradley JR, Daugherty LC, Delaneau O, Freson K, Garner SF, Grassi L, Guerrero J, Haimel M, Janssen-Megens EM, Kaan A, Kamat M, Kim B, Mandoli A, Marchini J, Martens JHA, Meacham S, Megy K, O’Connell J, Petersen R, Sharifi N, Sheard SM, Staley JR, Tuna S, van der Ent M, Walter K, Wang SY, Wheeler E, Wilder SP, Iotchkova V, Moore C, Sambrook J, Stunnenberg HG, Di Angelantonio E, Kaptoge S, Kuijpers TW, Carrillo-de-Santa-Pau E, Juan D, Rico D, Valencia A, Chen L, Ge B, Vasquez L, Kwan T, Garrido-Martin D, Watt S, Yang Y, Guigo R, Beck S, Paul DS, Pastinen T, Bujold D, Bourque G, Frontini M, Danesh J, Roberts DJ, Ouwehand WH, Butterworth AS, Soranzo N (2016) The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167(1415–1429):e19. 10.1016/j.cell.2016.10.042 - PMC - PubMed
-
- Bhat GR, Bhat A, Verma S, Sethi I, Shah R, Sharma V, Dar KA, Abrol D, Kaneez S, Kaul S, Ganju R, Kumar R (2019) Association of newly identified genetic variant rs2853677 of TERT with non-small cell lung cancer and leukemia in population of Jammu and Kashmir. India BMC Cancer 19:493. 10.1186/s12885-019-5685-2 - PMC - PubMed
-
- Chen MH, Raffield LM, Mousas A, Sakaue S, Huffman JE, Moscati A, Trivedi B, Jiang T, Akbari P, Vuckovic D, Bao EL, Zhong X, Manansala R, Laplante V, Chen M, Lo KS, Qian H, Lareau CA, Beaudoin M, Hunt KA, Akiyama M, Bartz TM, Ben-Shlomo Y, Beswick A, Bork-Jensen J, Bottinger EP, Brody JA, van Rooij FJA, Chitrala K, Cho K, Choquet H, Correa A, Danesh J, Di Angelantonio E, Dimou N, Ding J, Elliott P, Esko T, Evans MK, Floyd JS, Broer L, Grarup N, Guo MH, Greinacher A, Haessler J, Hansen T, Howson JMM, Huang QQ, Huang W, Jorgenson E, Kacprowski T, Kahonen M, Kamatani Y, Kanai M, Karthikeyan S, Koskeridis F, Lange LA, Lehtimaki T, Lerch MM, Linneberg A, Liu Y, Lyytikainen LP, Manichaikul A, Martin HC, Matsuda K, Mohlke KL, Mononen N, Murakami Y, Nadkarni GN, Nauck M, Nikus K, Ouwehand WH, Pankratz N, Pedersen O, Preuss M, Psaty BM, Raitakari OT, Roberts DJ, Rich SS, Rodriguez BAT, Rosen JD, Rotter JI, Schubert P, Spracklen CN, Surendran P, Tang H, Tardif JC, Trembath RC, Ghanbari M, Volker U, Volzke H, Watkins NA, Zonderman AB, Wilson PWF, Li Y, Butterworth AS, Gauchat JF, Chiang CWK, Li B, Loos RJF et al (2020) Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182(1198–1213):e14. 10.1016/j.cell.2020.06.045 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

