Predicting nicotine metabolism across ancestries using genotypes
- PMID: 36131240
- PMCID: PMC9490935
- DOI: 10.1186/s12864-022-08884-z
Predicting nicotine metabolism across ancestries using genotypes
Abstract
Background: There is a need to match characteristics of tobacco users with cessation treatments and risks of tobacco attributable diseases such as lung cancer. The rate in which the body metabolizes nicotine has proven an important predictor of these outcomes. Nicotine metabolism is primarily catalyzed by the enzyme cytochrone P450 (CYP2A6) and CYP2A6 activity can be measured as the ratio of two nicotine metabolites: trans-3'-hydroxycotinine to cotinine (NMR). Measurements of these metabolites are only possible in current tobacco users and vary by biofluid source, timing of collection, and protocols; unfortunately, this has limited their use in clinical practice. The NMR depends highly on genetic variation near CYP2A6 on chromosome 19 as well as ancestry, environmental, and other genetic factors. Thus, we aimed to develop prediction models of nicotine metabolism using genotypes and basic individual characteristics (age, gender, height, and weight).
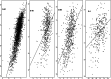
Results: We identified four multiethnic studies with nicotine metabolites and DNA samples. We constructed a 263 marker panel from filtering genome-wide association scans of the NMR in each study. We then applied seven machine learning techniques to train models of nicotine metabolism on the largest and most ancestrally diverse dataset (N=2239). The models were then validated using the other three studies (total N=1415). Using cross-validation, we found the correlations between the observed and predicted NMR ranged from 0.69 to 0.97 depending on the model. When predictions were averaged in an ensemble model, the correlation was 0.81. The ensemble model generalizes well in the validation studies across ancestries, despite differences in the measurements of NMR between studies, with correlations of: 0.52 for African ancestry, 0.61 for Asian ancestry, and 0.46 for European ancestry. The most influential predictors of NMR identified in more than two models were rs56113850, rs11878604, and 21 other genetic variants near CYP2A6 as well as age and ancestry.
Conclusions: We have developed an ensemble of seven models for predicting the NMR across ancestries from genotypes and age, gender and BMI. These models were validated using three datasets and associate with nicotine dosages. The knowledge of how an individual metabolizes nicotine could be used to help select the optimal path to reducing or quitting tobacco use, as well as, evaluating risks of tobacco use.
Keywords: Machine learning; Nicotine biomarkers; Nicotine metabolism; Polygenic risk score; Smoking cessation; Statistical learning.
© 2022. The Author(s).
Conflict of interest statement
JWB and CME are members and employees of BioRealm LLC. AWB is an employee of Oregon Research Institute and ORI Community and Evaluation Services, and serves as a Scientific Advisor and Consultant to BioRealm LLC. JWB, CSM and AWB are co-inventors on a related patent application “Biosignature Discovery for Substance Use Disorder Using Statistical Learning”, assigned to BioRealm LLC. BioRealm LLC offers genotyping and data analysis services. Other authors declare that they have no competing interests.
Figures

References
-
- Roberts W, Marotta PL, Verplaetse TL, Peltier MR, Burke C, Ramchandani VA, et al. A prospective study of the association between rate of nicotine metabolism and alcohol use in tobacco users in the United States. Drug Alcohol Depend. 2020;216:108210. doi: 10.1016/j.drugalcdep.2020.108210. - DOI - PMC - PubMed
-
- Lerman C, Schnoll RA, Hawk LW, Jr, Cinciripini P, George TP, Wileyto EP, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: a randomised, double-blind placebo-controlled trial. Lancet Respir Med. 2015;3(2):131–8. doi: 10.1016/S2213-2600(14)70294-2. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

