Estimating the effective reproduction number for heterogeneous models using incidence data
- PMID: 36133147
- PMCID: PMC9449464
- DOI: 10.1098/rsos.220005
Estimating the effective reproduction number for heterogeneous models using incidence data
Abstract
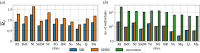
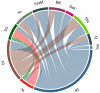
The effective reproduction number, , plays a key role in the study of infectious diseases, indicating the current average number of new infections caused by an infected individual in an epidemic process. Estimation methods for the time evolution of , using incidence data, rely on the generation interval distribution, g(τ), which is usually obtained from empirical data or theoretical studies using simple epidemic models. However, for systems that present heterogeneity, either on the host population or in the expression of the disease, there is a lack of data and of a suitable general methodology to obtain g(τ). In this work, we use mathematical models to bridge this gap. We present a general methodology for obtaining explicit expressions of the reproduction numbers and the generation interval distributions, within and between model sub-compartments provided by an arbitrary compartmental model. Additionally, we present the appropriate expressions to evaluate those reproduction numbers using incidence data. To highlight the relevance of such methodology, we apply it to the spread of COVID-19 in municipalities of the state of Rio de Janeiro, Brazil. Using two meta-population models, we estimate the reproduction numbers and the contributions of each municipality in the generation of cases in all others.
Keywords: COVID-19; effective reproduction number; mathematical models; meta-population models.
© 2022 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures


References
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous
