A nano-biomimetic transformation system enables in planta expression of a reporter gene in mature plants and seeds
- PMID: 36133668
- PMCID: PMC9417712
- DOI: 10.1039/d1na00107h
A nano-biomimetic transformation system enables in planta expression of a reporter gene in mature plants and seeds
Abstract
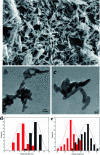
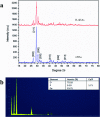
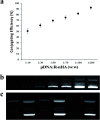
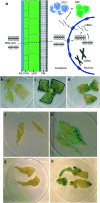
Plant genetic engineering will be essential to decipher the genomic basis of complex traits, optimize crop genomics, and enable plant-based production of recombinant proteins. However, established plant transformation approaches for bioengineering are fraught with limitations. Although nanoparticle-mediated methods show great promise for advancing plant biotechnology, many engineered nanomaterials can have cytotoxic and ecological effects. Here, we demonstrate the efficient uptake of a nano-biomimetic carrier of plasmid DNA and transient expression of a reporter gene in leaves of Arabidopsis, common ice plant and tobacco, as well as in the developing seed tissues of Arabidopsis, field mustard, barley, and wheat. The nano-biomimetic transformation system described here has all the advantages of other nanoparticle-mediated approaches for passive delivery of genetic cargo into a variety of plant species and is also nontoxic to cells and to the environment for diverse biotechnological applications in plant biology and crop science.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

Similar articles
-
Carbon Nanotube-Mediated Plasmid DNA Delivery in Rice Leaves and Seeds.Int J Mol Sci. 2022 Apr 7;23(8):4081. doi: 10.3390/ijms23084081. Int J Mol Sci. 2022. PMID: 35456898 Free PMC article.
-
Enhancement of Plant Productivity in the Post-Genomics Era.Curr Genomics. 2016 Aug;17(4):295-6. doi: 10.2174/138920291704160607182507. Curr Genomics. 2016. PMID: 27499678 Free PMC article.
-
High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants.Nat Nanotechnol. 2019 May;14(5):456-464. doi: 10.1038/s41565-019-0382-5. Epub 2019 Feb 25. Nat Nanotechnol. 2019. PMID: 30804481 Free PMC article.
-
Nanotechnology Approaches for Chloroplast Biotechnology Advancements.Front Plant Sci. 2021 Jul 26;12:691295. doi: 10.3389/fpls.2021.691295. eCollection 2021. Front Plant Sci. 2021. PMID: 34381480 Free PMC article. Review.
-
The Promising Nanovectors for Gene Delivery in Plant Genome Engineering.Int J Mol Sci. 2022 Jul 31;23(15):8501. doi: 10.3390/ijms23158501. Int J Mol Sci. 2022. PMID: 35955636 Free PMC article. Review.
Cited by
-
Application of Nanotechnology in Plant Genetic Engineering.Int J Mol Sci. 2023 Oct 2;24(19):14836. doi: 10.3390/ijms241914836. Int J Mol Sci. 2023. PMID: 37834283 Free PMC article. Review.
-
DNA delivery into plant tissues using carbon dots made from citric acid and β-alanine.Front Chem. 2025 Mar 19;13:1542504. doi: 10.3389/fchem.2025.1542504. eCollection 2025. Front Chem. 2025. PMID: 40177349 Free PMC article.
-
An efficient and broadly applicable method for transient transformation of plants using vertically aligned carbon nanofiber arrays.Front Plant Sci. 2022 Nov 23;13:1051340. doi: 10.3389/fpls.2022.1051340. eCollection 2022. Front Plant Sci. 2022. PMID: 36507425 Free PMC article.
-
Acid-Triggered Release of Eugenol and Fluoride by Desensitizing Macro- and Nanoparticles.J Funct Biomater. 2023 Jan 11;14(1):42. doi: 10.3390/jfb14010042. J Funct Biomater. 2023. PMID: 36662089 Free PMC article.
-
Nano-enabled agriculture: How do nanoparticles cross barriers in plants?Plant Commun. 2022 Nov 14;3(6):100346. doi: 10.1016/j.xplc.2022.100346. Epub 2022 Jun 9. Plant Commun. 2022. PMID: 35689377 Free PMC article. Review.
References
-
- Altpeter F. Springer N. M. Bartley L. E. Blechl A. E. Brutnell T. P. Citovsky V. Conrad L. J. Gelvin S. B. Jackson D. P. Kausch A. P. Lemaux P. G. Medford J. I. Orozco-Cárdenas M. L. Tricoli D. M. Van Eck J. Voytas D. F. Walbot V. Wang K. Zhang Z. J. Stewart C. N. Plant Cell. 2016;28:1510–1520. - PMC - PubMed
LinkOut - more resources
Full Text Sources

