Non-canonical DNA structures: Diversity and disease association
- PMID: 36134025
- PMCID: PMC9483843
- DOI: 10.3389/fgene.2022.959258
Non-canonical DNA structures: Diversity and disease association
Abstract
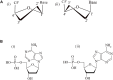
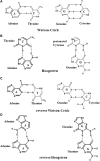
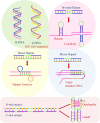
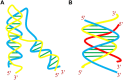
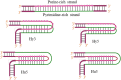
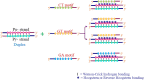
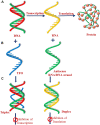
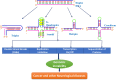
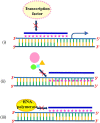
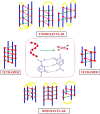
A complete understanding of DNA double-helical structure discovered by James Watson and Francis Crick in 1953, unveil the importance and significance of DNA. For the last seven decades, this has been a leading light in the course of the development of modern biology and biomedical science. Apart from the predominant B-form, experimental shreds of evidence have revealed the existence of a sequence-dependent structural diversity, unusual non-canonical structures like hairpin, cruciform, Z-DNA, multistranded structures such as DNA triplex, G-quadruplex, i-motif forms, etc. The diversity in the DNA structure depends on various factors such as base sequence, ions, superhelical stress, and ligands. In response to these various factors, the polymorphism of DNA regulates various genes via different processes like replication, transcription, translation, and recombination. However, altered levels of gene expression are associated with many human genetic diseases including neurological disorders and cancer. These non-B-DNA structures are expected to play a key role in determining genetic stability, DNA damage and repair etc. The present review is a modest attempt to summarize the available literature, illustrating the occurrence of non-canonical structures at the molecular level in response to the environment and interaction with ligands and proteins. This would provide an insight to understand the biological functions of these unusual DNA structures and their recognition as potential therapeutic targets for diverse genetic diseases.
Keywords: G-quadruplex; Z-DNA; cruciform; non-canonical DNA; triplex.
Copyright © 2022 Bansal, Kaushik and Kukreti.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Adachi M., Tsujimoto Y. (1990). Potential Z-DNA elements surround the breakpoints of chromosome translocation within the 5'flanking region of bcl-2 gene. Oncogene 5 (11), 1653–1657. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

