Transcriptome-Wide Analysis Revealed the Potential of the High-Affinity Potassium Transporter (HKT) Gene Family in Rice Salinity Tolerance via Ion Homeostasis
- PMID: 36134956
- PMCID: PMC9495969
- DOI: 10.3390/bioengineering9090410
Transcriptome-Wide Analysis Revealed the Potential of the High-Affinity Potassium Transporter (HKT) Gene Family in Rice Salinity Tolerance via Ion Homeostasis
Abstract
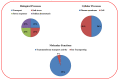
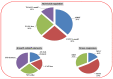
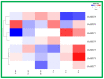
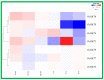
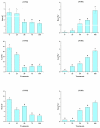
The high-affinity potassium transporter (HKT) genes are key ions transporters, regulating the plant response to salt stress via sodium (Na+) and potassium (K+) homeostasis. The main goal of this research was to find and understand the HKT genes in rice and their potential biological activities in response to brassinosteroids (BRs), jasmonic acid (JA), seawater, and NaCl stress. The in silico analyses of seven OsHKT genes involved their evolutionary tree, gene structures, conserved motifs, and chemical properties, highlighting the key aspects of OsHKT genes. The Gene Ontology (GO) analysis of HKT genes revealed their roles in growth and stress responses. Promoter analysis showed that the majority of the HKT genes participate in abiotic stress responses. Tissue-specific expression analysis showed higher transcriptional activity of OsHKT genes in roots and leaves. Under NaCl, BR, and JA application, OsHKT1 was expressed differentially in roots and shoots. Similarly, the induced expression pattern of OsHKT1 was recorded in the seawater resistant (SWR) cultivar. Additionally, the Na+ to K+ ratio under different concentrations of NaCl stress has been evaluated. Our data highlighted the important role of the OsHKT gene family in regulating the JA and BR mediated rice salinity tolerance and could be useful for rice future breeding programs.
Keywords: HKT; hormones; rice; salinity; seawater.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures








References
-
- Ammiraju J.S., Luo M., Goicoechea J.L., Wang W., Kudrna D., Mueller C., Talag J., Kim H., Sisneros N.B., Blackmon B., et al. The Oryza bacterial artificial chromosome library resource: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 2006;16:140–147. doi: 10.1101/gr.3766306. - DOI - PMC - PubMed
Grants and funding
- 32101817/National Natural Science Foundation of China
- (CX(21)3111/Jiangsu Agricultural Science and Technology Innovation Fund
- 21KJD210001/Natural Science Foundation of the Jiangsu Higher Education Institutions
- BE2022304/Scientific and Technological Innovation Fund of Carbon Emissions Peak and Neutrality of Jiangsu Provincial Department of Science and Technology
LinkOut - more resources
Full Text Sources

