Mitochondrial DNA barcoding of mosquito species (Diptera: Culicidae) in Thailand
- PMID: 36137118
- PMCID: PMC9642330
- DOI: 10.1371/journal.pone.0275090
Mitochondrial DNA barcoding of mosquito species (Diptera: Culicidae) in Thailand
Erratum in
-
Correction: Mitochondrial DNA barcoding of mosquito species (Diptera: Culicidae) in Thailand.PLoS One. 2025 Jan 3;20(1):e0317172. doi: 10.1371/journal.pone.0317172. eCollection 2025. PLoS One. 2025. PMID: 39752373 Free PMC article.
Abstract
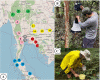
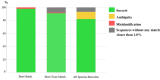
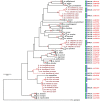
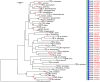
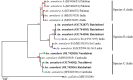
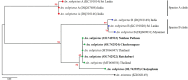
The correct identification of mosquito species is important for effective mosquito vector control. However, the standard morphological identification of mosquito species based on the available keys is not easy with specimens in the field due to missing or damaged morphological features during mosquito collections, often leading to the misidentification of morphologically indistinguishable. To resolve this problem, we collected mosquito species across Thailand to gather genetic information, and evaluated the DNA barcoding efficacy for mosquito species identification in Thailand. A total of 310 mosquito samples, representing 73 mosquito species, were amplified using mitochondrial cytochrome c oxidase subunit I (COI) primers. The average maximum intraspecific genetic variation of the 73 mosquito species was 1% ranged from 0-5.7%. While, average minimum interspecific genetic variation (the distance to the nearest neighbour) of the 73 mosquito species was 7% ranged from 0.3-12.9%. The identification of success rates based on the "Best Match," "Best Close Match," and "All Species Barcodes" methods were 97.7%, 91.6%, and 81%, respectively. Phylogenetic analyses of Anopheles COI sequences demonstrated a clear separation between almost all species (except for those between An. baimaii and An. dirus), with high bootstrap support values (97%-99%). Furthermore, phylogenetic analyses revealed potential sibling species of An. annularis, An. tessellatus, and An. subpictus in Thailand. Our results indicated that DNA barcoding is an effective molecular approach for the accurate identification of mosquitoes in Thailand.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Molecular characterization of Anopheline (Diptera: Culicidae) mosquitoes from eight geographical locations of Sri Lanka.Malar J. 2017 Jun 2;16(1):234. doi: 10.1186/s12936-017-1876-y. Malar J. 2017. PMID: 28578667 Free PMC article.
-
DNA barcoding of morphologically characterized mosquitoes belonging to the subfamily Culicinae from Sri Lanka.Parasit Vectors. 2018 Apr 25;11(1):266. doi: 10.1186/s13071-018-2810-z. Parasit Vectors. 2018. PMID: 29695263 Free PMC article.
-
Island mosquitoes of Thailand: an update on species diversity and DNA barcoding.Parasitol Res. 2024 May 29;123(5):224. doi: 10.1007/s00436-024-08237-7. Parasitol Res. 2024. PMID: 38809447
-
Identification of Belgian mosquito species (Diptera: Culicidae) by DNA barcoding.Mol Ecol Resour. 2015 Mar;15(2):449-57. doi: 10.1111/1755-0998.12318. Epub 2014 Sep 15. Mol Ecol Resour. 2015. PMID: 25143182
-
DNA barcoding mosquitoes: advice for potential prospectors.Parasitology. 2018 Apr;145(5):622-633. doi: 10.1017/S0031182018000343. Epub 2018 Mar 22. Parasitology. 2018. PMID: 29564995 Review.
Cited by
-
Real-time dynamic monitoring and multiplex PCR identification of vector mosquitoes in Zhejiang, China.PLoS Negl Trop Dis. 2025 May 27;19(5):e0013129. doi: 10.1371/journal.pntd.0013129. eCollection 2025 May. PLoS Negl Trop Dis. 2025. PMID: 40424257 Free PMC article.
-
Discobola Osten Sacken, 1865 (Diptera, Limoniidae) in China: Taxonomic Review, Updated Distribution, and DNA Barcoding.Insects. 2025 Aug 15;16(8):845. doi: 10.3390/insects16080845. Insects. 2025. PMID: 40870646 Free PMC article.
-
Geometric morphometric and molecular techniques for discriminating among three cryptic species of the Anopheles barbirostris complex (diptera: culicidae) in Thailand.Heliyon. 2022 Oct 26;8(10):e11261. doi: 10.1016/j.heliyon.2022.e11261. eCollection 2022 Oct. Heliyon. 2022. PMID: 36339998 Free PMC article.
-
Correction: Mitochondrial DNA barcoding of mosquito species (Diptera: Culicidae) in Thailand.PLoS One. 2025 Jan 3;20(1):e0317172. doi: 10.1371/journal.pone.0317172. eCollection 2025. PLoS One. 2025. PMID: 39752373 Free PMC article.
-
Evaluation of Modern Techniques for Species Identification of Lutzia Mosquitoes (Diptera: Culicidae) in Thailand: Geometric Morphometrics and DNA Barcoding.Insects. 2023 Jan 12;14(1):78. doi: 10.3390/insects14010078. Insects. 2023. PMID: 36662006 Free PMC article.
References
-
- Harbach RE. Mosquito Taxonomic Inventory. 2022. Available from: https://mosquito-taxonomic-inventory.myspecies.info.
-
- Killick-Kendrick R. Medical entomology for students. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1996. 10.1016/S0035-9203(96)90345-4. - DOI
-
- World Health Organization. WHO Factsheet Vector-borne diseases. Factsheet number 387. 2014; 10. Available from: http://www.who.int/kobe_centre/mediacentre/ vbdfact sheet.pdf.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

