Patient-derived monoclonal antibody neutralizes HCV infection in vitro and vivo without generating escape mutants
- PMID: 36137152
- PMCID: PMC9499215
- DOI: 10.1371/journal.pone.0274283
Patient-derived monoclonal antibody neutralizes HCV infection in vitro and vivo without generating escape mutants
Abstract
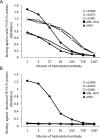
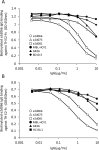
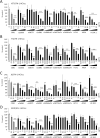
In recent years, new direct-acting antivirals for hepatitis C virus (HCV) have been approved, but hepatitis C continues to pose a threat to human health. It is important to develop neutralizing anti-HCV antibodies to prevent medical and accidental infection, such as might occur via liver transplantation of chronic HCV patients and needle-stick accidents in the clinic. In this study, we sought to obtain anti-HCV antibodies using phage display screening. Phages displaying human hepatocellular carcinoma patient-derived antibodies were screened by 4 rounds of biopanning with genotype-1b and -2a HCV envelope E2 protein adsorbed to magnetic beads. The three antibodies obtained from this screen had reactivity against E2 proteins derived from both genotype-1b and -2a strains. However, in epitope analysis, these antibodies did not recognize linear peptides from an overlapping E2 epitope peptide library, and did not bind to denatured E2 protein. In addition, these antibodies showed cross-genotypic neutralizing activity against genotype-1a, -1b, -2a, and -3a cell culture-generated infectious HCV particles (HCVcc). Moreover, emergence of viral escape mutants was not observed after repeated rounds of passaging of HCV-infected cells in the presence of one such antibody, e2d066. Furthermore, injection of the e2d066 antibody into human hepatocyte-transplanted immunodeficient mice inhibited infection by J6/JFH-1 HCVcc. In conclusion, we identified conformational epitope-recognizing, cross-genotypic neutralizing antibodies using phage display screening. Notably, e2d066 antibody did not select for escape mutant emergence in vitro and demonstrated neutralizing activity in vivo. Our results suggested that these antibodies may serve as prophylactic and therapeutic agents.
Conflict of interest statement
Hiroshi Yokokawa, Noriko Nakamura, Tomokatsu Iwamura, and Hideki Narumi are employees of Toray Industries, Inc. Midori Shinohara is an employee of Medical & Biological Laboratories Co., Ltd. The researchers were employed by these commercial funders and received funding for patent rights to this research. This study was patent filed in 2015 (WO2015/141826), but the rights application was withdrawn in 2018. The researchers and commercial funders do not benefit from the submission of this study to PLOS ONE. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures
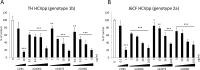

References
-
- Hepatitis C fact sheet. World Health Organization. July 2021. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

