Shared Clavulanate and Tazobactam Antigenic Determinants Activate T-Cells from Hypersensitive Patients
- PMID: 36137197
- PMCID: PMC9682523
- DOI: 10.1021/acs.chemrestox.2c00231
Shared Clavulanate and Tazobactam Antigenic Determinants Activate T-Cells from Hypersensitive Patients
Abstract
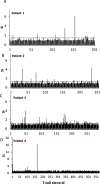
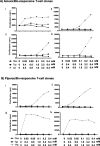
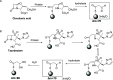
β-Lactamase inhibitors such as clavulanic acid and tazobactam were developed to overcome β-lactam antibiotic resistance. Hypersensitivity reactions to these drugs have not been studied in detail, and the antigenic determinants that activate T-cells have not been defined. The objectives of this study were to (i) characterize clavulanate- and tazobactam-responsive T-cells from hypersensitive patients, (ii) explore clavulanate and tazobactam T-cell crossreactivity, and (iii) define the antigenic determinants that contribute to T-cell reactivity. Antigen specificity, pathways of T-cell activation, and crossreactivity with clavulanate- and tazobactam-specific T-cell clones were assessed by proliferation and cytokine release assays. Antigenic determinants were analyzed by mass spectrometry-based proteomics methods. Clavulanate- and tazobactam-responsive CD4+ T-cell clones were stimulated to proliferate and secrete IFN-γ in an MHC class II-restricted and dose-dependent manner. T-cell activation with clavulanate- and tazobactam was dependent on antigen presenting cells because their fixation prevented the T-cell response. Strong crossreactivity was observed between clavulanate- and tazobactam-T-cells; however, neither drug activated β-lactam antibiotic-responsive T-cells. Mass spectrometric analysis revealed that both compounds form multiple antigenic determinants with lysine residues on proteins, including an overlapping aldehyde and hydrated aldehyde adduct with mass additions of 70 and 88 Da, respectively. Collectively, these data show that although clavulanate and tazobactam are structurally distinct, the antigenic determinants formed by both drugs overlap, which explains the observed T-cell cross-reactivity.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Gavin P. J.; Suseno M. T.; Thomson R. B. Jr.; Gaydos J. M.; Pierson C. L.; Halstead D. C.; Aslanzadeh J.; Brecher S.; Rotstein C.; Brossette S. E.; Peterson L. R. Clinical correlation of the CLSI susceptibility breakpoint for piperacillin- tazobactam against extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella species. Antimicrob. Agents Chemother. 2006, 50, 2244–2247. 10.1128/AAC.00381-05. - DOI - PMC - PubMed
-
- Jones R. N.; Pfaller M. A.; Fuchs P. C.; Aldridge K.; Allen S. D.; Gerlach E. H. Piperacillin/tazobactam (YTR 830) combination. Comparative antimicrobial activity against 5889 recent aerobic clinical isolates and 60 Bacteroides fragilis group strains. Diagn. Microbiol. Infect. Dis. 1989, 12, 489–494. 10.1016/0732-8893(89)90083-7. - DOI - PubMed

