A fluorescent sensor for real-time measurement of extracellular oxytocin dynamics in the brain
- PMID: 36138174
- PMCID: PMC9550624
- DOI: 10.1038/s41592-022-01597-x
A fluorescent sensor for real-time measurement of extracellular oxytocin dynamics in the brain
Abstract
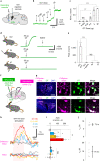
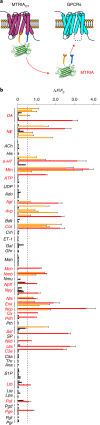
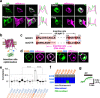
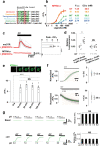
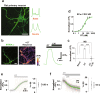
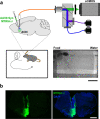
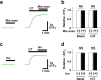
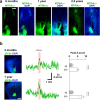
Oxytocin (OT), a hypothalamic neuropeptide that acts as a neuromodulator in the brain, orchestrates a variety of animal behaviors. However, the relationship between brain OT dynamics and complex animal behaviors remains largely elusive, partly because of the lack of a suitable technique for its real-time recording in vivo. Here, we describe MTRIAOT, a G-protein-coupled receptor-based green fluorescent OT sensor that has a large dynamic range, suitable affinity, ligand specificity for OT orthologs, minimal effects on downstream signaling and long-term fluorescence stability. By combining viral gene delivery and fiber photometry-mediated fluorescence measurements, we demonstrate the utility of MTRIAOT for real-time detection of brain OT dynamics in living mice. MTRIAOT-mediated measurements indicate variability of OT dynamics depending on the behavioral context and physical condition of an animal. MTRIAOT will likely enable the analysis of OT dynamics in a variety of physiological and pathological processes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

