Cellular and transcriptional dynamics of human neutrophils at steady state and upon stress
- PMID: 36138183
- PMCID: PMC7615267
- DOI: 10.1038/s41590-022-01311-1
Cellular and transcriptional dynamics of human neutrophils at steady state and upon stress
Abstract
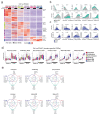
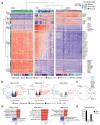
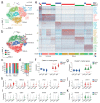
Traditionally viewed as poorly plastic, neutrophils are now recognized as functionally diverse; however, the extent and determinants of neutrophil heterogeneity in humans remain unclear. We performed a comprehensive immunophenotypic and transcriptome analysis, at a bulk and single-cell level, of neutrophils from healthy donors and patients undergoing stress myelopoiesis upon exposure to growth factors, transplantation of hematopoietic stem cells (HSC-T), development of pancreatic cancer and viral infection. We uncover an extreme diversity of human neutrophils in vivo, reflecting the rates of cell mobilization, differentiation and exposure to environmental signals. Integrated control of developmental and inducible transcriptional programs linked flexible granulopoietic outputs with elicitation of stimulus-specific functional responses. In this context, we detected an acute interferon (IFN) response in the blood of patients receiving HSC-T that was mirrored by marked upregulation of IFN-stimulated genes in neutrophils but not in monocytes. Systematic characterization of human neutrophil plasticity may uncover clinically relevant biomarkers and support the development of diagnostic and therapeutic tools.
© 2022. Springer Nature America, Inc.
Conflict of interest statement
The authors declare no competing interests
Figures















References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

