Transcriptome Analyses Identify Deregulated MYC in Early Onset Colorectal Cancer
- PMID: 36139061
- PMCID: PMC9496520
- DOI: 10.3390/biom12091223
Transcriptome Analyses Identify Deregulated MYC in Early Onset Colorectal Cancer
Abstract
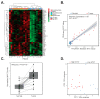
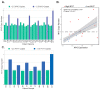
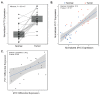
Despite a global decrease in colorectal cancer (CRC) incidence, the prevalence of early-onset colorectal cancer (EOCRC), or those occurring in individuals before the age of 50, has steadily increased over the past several decades. When compared to later onset colorectal cancer (LOCRC) in individuals over 50, our understanding of the genetic and molecular underpinnings of EOCRCs is limited. Here, we conducted transcriptomic analyses of patient-matched normal colonic segments and tumors to identify gene expression programs involved in carcinogenesis. Amongst differentially expressed genes, we found increased expression of the c-MYC proto-oncogene (MYC) and its downstream targets in tumor samples. We identified tumors with high and low differential MYC expression and found patients with high-MYC tumors were older and overweight or obese. We also detected elevated expression of the PVT1 long-non-coding RNA (lncRNA) in most tumors and found gains in copy number for both MYC and PVT1 gene loci in 35% of tumors evaluated. Our transcriptome analyses indicate that EOCRC can be sub-classified into groups based on differential MYC expression and suggest that deregulated MYC contributes to CRCs that develop in younger patients.
Keywords: MYC; PVT1; RNA-sequencing; early-onset colorectal cancer; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Cercek A., Chatila W.K., Yaeger R., Walch H., Fernandes G.D.S., Krishnan A., Palmaira L., Maio A., Kemel Y., Srinivasan P., et al. A Comprehensive Comparison of Early-Onset and Average-Onset Colorectal Cancers. JNCI J. Natl. Cancer Inst. 2021;113:1683–1692. doi: 10.1093/jnci/djab124. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

