Comparative Transcriptomic Analysis of mRNAs, miRNAs and lncRNAs in the Longissimus dorsi Muscles between Fat-Type and Lean-Type Pigs
- PMID: 36139132
- PMCID: PMC9496231
- DOI: 10.3390/biom12091294
Comparative Transcriptomic Analysis of mRNAs, miRNAs and lncRNAs in the Longissimus dorsi Muscles between Fat-Type and Lean-Type Pigs
Abstract
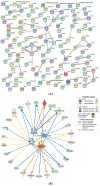
In pigs, meat quality and production are two important traits affecting the pig industry and human health. Compared to lean-type pigs, fat-type pigs contain higher intramuscular fat (IMF) contents, better taste and nutritional value. To uncover genetic factors controlling differences related to IMF in pig muscle, we performed RNA-seq analysis on the transcriptomes of the Longissimus dorsi (LD) muscle of Laiwu pigs (LW, fat-type pigs) and commercial Duroc × Landrace × Yorkshire pigs (DLY, lean-type pigs) at 150 d to compare the expression profiles of mRNA, miRNA and lncRNA. A total of 225 mRNAs, 12 miRNAs and 57 lncRNAs were found to be differentially expressed at the criteria of |log2(foldchange)| > 1 and q < 0.05. The mRNA expression of LDHB was significantly higher in the LD muscle of LW compared to DLY pigs with log2(foldchange) being 9.66. Using protein interaction prediction method, we identified more interactions of estrogen-related receptor alpha (ESRRA) associated with upregulated mRNAs, whereas versican (VCAN) and proenkephalin (PENK) were associated with downregulated mRNAs in LW pigs. Integrated analysis on differentially expressed (DE) mRNAs and miRNAs in the LD muscle between LW and DLY pigs revealed two network modules: between five upregulated mRNA genes (GALNT15, FKBP5, PPARGC1A, LOC110258214 and LOC110258215) and six downregulated miRNA genes (ssc-let-7a, ssc-miR190-3p, ssc-miR356-5p, ssc-miR573-5p, ssc-miR204-5p and ssc-miR-10383), and between three downregulated DE mRNA genes (IFRD1, LOC110258600 and LOC102158401) and six upregulated DE miRNA genes (ssc-miR1379-3p, ssc-miR1379-5p, ssc-miR397-5p, ssc-miR1358-5p, ssc-miR299-5p and ssc-miR1156-5p) in LW pigs. Based on the mRNA and ncRNA binding site targeting database, we constructed a regulatory network with miRNA as the center and mRNA and lncRNA as the target genes, including GALNT15/ssc-let-7a/LOC100523888, IFRD1/ssc-miR1379-5p/CD99, etc., forming a ceRNA network in the LD muscles that are differentially expressed between LW and DLY pigs. Collectively, these data may provide resources for further investigation of molecular mechanisms underlying differences in meat traits between lean- and fat-type pigs.
Keywords: Longissimus dorsi muscle; intramuscular fat; lncRNA; mRNA; miRNA; pig; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

