Proteomic Analysis of Retinal Mitochondria-Associated ER Membranes Identified Novel Proteins of Retinal Degeneration in Long-Term Diabetes
- PMID: 36139394
- PMCID: PMC9497316
- DOI: 10.3390/cells11182819
Proteomic Analysis of Retinal Mitochondria-Associated ER Membranes Identified Novel Proteins of Retinal Degeneration in Long-Term Diabetes
Abstract
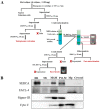
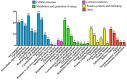
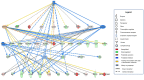
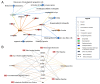
The mitochondria-associated endoplasmic reticulum (ER) membrane (MAM) is the physical contact site between the ER and the mitochondria and plays a vital role in the regulation of calcium signaling, bioenergetics, and inflammation. Disturbances in these processes and dysregulation of the ER and mitochondrial homeostasis contribute to the pathogenesis of diabetic retinopathy (DR). However, few studies have examined the impact of diabetes on the retinal MAM and its implication in DR pathogenesis. In the present study, we investigated the proteomic changes in retinal MAM from Long Evans rats with streptozotocin-induced long-term Type 1 diabetes. Furthermore, we performed in-depth bioinformatic analysis to identify key MAM proteins and pathways that are potentially implicated in retinal inflammation, angiogenesis, and neurodegeneration. A total of 2664 unique proteins were quantified using IonStar proteomics-pipeline in rat retinal MAM, among which 179 proteins showed significant changes in diabetes. Functional annotation revealed that the 179 proteins are involved in important biological processes such as cell survival, inflammatory response, and cellular maintenance, as well as multiple disease-relevant signaling pathways, e.g., integrin signaling, leukocyte extravasation, PPAR, PTEN, and RhoGDI signaling. Our study provides comprehensive information on MAM protein changes in diabetic retinas, which is helpful for understanding the mechanisms of metabolic dysfunction and retinal cell injury in DR.
Keywords: Type 1 diabetes; diabetic retinopathy; mitochondria-associated ER membrane; proteomic; retinal degeneration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Reis A., Mateus C., Melo P., Figueira J., Cunha-Vaz J., Castelo-Branco M. Pure neuroretinal dysfunction in diabetic retinopathy occurring prior to endothelial and vascular damage. Acta Ophthalmol. 2013;91:S252. doi: 10.1111/j.1755-3768.2013.1662.x. - DOI
-
- Lee V.K., Hosking B.M., Holeniewska J., Kubala E.C., Lundh von Leithner P., Gardner P.J., Foxton R.H., Shima D.T. BTBR ob/ob mouse model of type 2 diabetes exhibits early loss of retinal function and retinal inflammation followed by late vascular changes. Diabetologia. 2018;61:2422–2432. doi: 10.1007/s00125-018-4696-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

