Androgen Receptor Splice Variants Contribute to the Upregulation of DNA Repair in Prostate Cancer
- PMID: 36139600
- PMCID: PMC9496991
- DOI: 10.3390/cancers14184441
Androgen Receptor Splice Variants Contribute to the Upregulation of DNA Repair in Prostate Cancer
Abstract
Background: Canonical androgen receptor (AR) signaling regulates a network of DNA repair genes in prostate cancer (PCA). Experimental and clinical evidence indicates that androgen deprivation not only suppresses DNA repair activity but is often synthetically lethal in combination with PARP inhibition. The present study aimed to elucidate the impact of AR splice variants (AR-Vs), occurring in advanced or late-stage PCA, on DNA repair machinery.
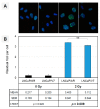
Methods: Two hundred and seventy-three tissue samples were analyzed, including primary hormone-naïve PCA, primary metastases, hormone-sensitive PCA on androgen deprivation therapy (ADT) and castration refractory PCA (CRPC group). The transcript levels of the target genes were profiled using the nCounter platform. Experimental support for the findings was gained in AR/AR-V7-expressing LNCaP cells subjected to ionizing radiation.
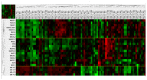
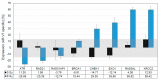
Results: AR-Vs were present in half of hormone-sensitive PCAs on androgen deprivation therapy (ADT) and two-thirds of CRPC samples. The presence of AR-Vs is highly correlated with increased activity in the AR pathway and DNA repair gene expression. In AR-V-expressing CRPC, the DNA repair score increased by 2.5-fold as compared to AR-V-negative samples. Enhanced DNA repair and the deregulation of DNA repair genes by AR-V7 supported the clinical data in a cell line model.
Conclusions: The expression of AR splice variants such as AR-V7 in PCA patients following ADT might be a reason for reduced or absent therapy effects in patients on additional PARP inhibition due to the modulation of DNA repair gene expression. Consequently, AR-Vs should be further studied as predictive biomarkers for therapy response in this setting.
Keywords: AR-V7; BRCA1; BRCA2; DNA repair; androgen deprivation therapy; androgen receptor; castration-refractory prostate cancer; prostate cancer; splice variant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
The positive relationship between androgen receptor splice variant-7 expression and the risk of castration-resistant prostate cancer: A cumulative analysis.Front Oncol. 2023 Feb 14;13:1053111. doi: 10.3389/fonc.2023.1053111. eCollection 2023. Front Oncol. 2023. PMID: 36865799 Free PMC article.
-
Ratio of the expression levels of androgen receptor splice variant 7 to androgen receptor in castration refractory prostate cancer.Oncol Lett. 2021 Dec;22(6):831. doi: 10.3892/ol.2021.13092. Epub 2021 Oct 13. Oncol Lett. 2021. PMID: 34691258 Free PMC article.
-
Transcript Levels of Androgen Receptor Variant 7 and Ubiquitin-Conjugating Enzyme 2C in Hormone Sensitive Prostate Cancer and Castration-Resistant Prostate Cancer.Prostate. 2017 Jan;77(1):60-71. doi: 10.1002/pros.23248. Epub 2016 Aug 22. Prostate. 2017. PMID: 27550197
-
Restoration of FKBP51 protein promotes the progression of castration resistant prostate cancer.Ann Transl Med. 2019 Dec;7(23):729. doi: 10.21037/atm.2019.11.127. Ann Transl Med. 2019. PMID: 32042745 Free PMC article.
-
Androgen receptors in hormone-dependent and castration-resistant prostate cancer.Pharmacol Ther. 2013 Dec;140(3):223-38. doi: 10.1016/j.pharmthera.2013.07.003. Epub 2013 Jul 13. Pharmacol Ther. 2013. PMID: 23859952 Review.
Cited by
-
Cell-Free DNA Sequencing Reveals Gene Variants in DNA Damage Repair Genes Associated with Prognosis of Prostate Cancer Patients.Cells. 2022 Nov 15;11(22):3618. doi: 10.3390/cells11223618. Cells. 2022. PMID: 36429046 Free PMC article.
-
The positive relationship between androgen receptor splice variant-7 expression and the risk of castration-resistant prostate cancer: A cumulative analysis.Front Oncol. 2023 Feb 14;13:1053111. doi: 10.3389/fonc.2023.1053111. eCollection 2023. Front Oncol. 2023. PMID: 36865799 Free PMC article.
-
Targeting the CLK2/SRSF9 splicing axis in prostate cancer leads to decreased ARV7 expression.Mol Oncol. 2025 Feb;19(2):496-518. doi: 10.1002/1878-0261.13728. Epub 2024 Sep 11. Mol Oncol. 2025. PMID: 39258426 Free PMC article.
-
Exploiting the DNA Damage Response for Prostate Cancer Therapy.Cancers (Basel). 2023 Dec 23;16(1):83. doi: 10.3390/cancers16010083. Cancers (Basel). 2023. PMID: 38201511 Free PMC article. Review.
-
The catalytic subunit of DNA-PK regulates transcription and splicing of AR in advanced prostate cancer.J Clin Invest. 2023 Nov 15;133(22):e169200. doi: 10.1172/JCI169200. J Clin Invest. 2023. PMID: 37751307 Free PMC article.
References
-
- Polkinghorn W.R., Parker J.S., Lee M.X., Kass E.M., Spratt D.E., Iaquinta P.J., Arora V.K., Yen W.F., Cai L., Zheng D., et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3:1245–1253. doi: 10.1158/2159-8290.CD-13-0172. - DOI - PMC - PubMed
-
- Goodwin J.F., Schiewer M.J., Dean J.L., Schrecengost R.S., de Leeuw R., Han S., Ma T., Den R.B., Dicker A.P., Feng F.Y., et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 2013;3:1254–1271. doi: 10.1158/2159-8290.CD-13-0108. - DOI - PMC - PubMed
-
- Li L., Karanika S., Yang G., Wang J., Park S., Broom B.M., Manyam G.C., Wu W., Luo Y., Basourakos S., et al. Androgen receptor inhibitor-induced ‘BRCAness’ and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci. Signal. 2017;10:eaam7479. doi: 10.1126/scisignal.aam7479. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

