Direct Comparison of HPV16 Viral Genomic Integration, Copy Loss, and Structural Variants in Oropharyngeal and Uterine Cervical Cancers Reveal Distinct Relationships to E2 Disruption and Somatic Alteration
- PMID: 36139648
- PMCID: PMC9496734
- DOI: 10.3390/cancers14184488
Direct Comparison of HPV16 Viral Genomic Integration, Copy Loss, and Structural Variants in Oropharyngeal and Uterine Cervical Cancers Reveal Distinct Relationships to E2 Disruption and Somatic Alteration
Abstract
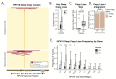
Squamous cell carcinoma of the oropharynx caused by HPV type 16 (HPV16+ OPSCC) is the most common HPV-associated malignancy in the USA and has many molecular differences from uterine cervical squamous cell carcinoma (UCSCC). Our understanding of HPV oncogenesis relied on studies of UCSCC revealing a consensus model reliant on HPV integration with a loss of E2. Here, we compare patterns of HPV integration in UCSCC and OPSCC by analysis of affinity capture sequencing of the HPV16 genome in 104 OPSCC and 44 UCSCC tumors. These cohorts were contemporaneously sequenced using an identical strategy. Integration was identified using discordant read pair clustering and assembly-based approaches. Viral integration sites, structural variants, and copy losses were examined. While large-scale deep losses of HPV16 genes were common in UCSCC and were associated with E2 loss, deep copy losses of the HPV16 genome were infrequent in HPV16+ OPSCC. Similarly, structural variants within HPV16 favored E2 loss in UCSCC but not OPSCC. HPV16 integration sites were non-random, with recurrent integration hot-spots identified. OPSCC tumors had many more integration sites per tumor when compared to UCSCC and had more integration sites in genomic regions with high gene density. These data show that viral integration and E2 disruption are distinct in UCSCC and OPSCC. Our findings also add to growing literature suggesting that HPV tumorigenesis in OPSCC does not follow the model developed based on UCSCC.
Keywords: HPV16; integration; oropharynx; squamous cell carcinoma; uterine cervix.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Schiffman M.H., Bauer H.M., Hoover R.N., Glass A.G., Cadell D.M., Rush B.B., Scott D.R., Sherman M.E., Kurman R.J., Wacholder S. Epidemiologic Evidence Showing That Human Papillomavirus Infection Causes Most Cervical Intraepithelial Neoplasia. J. Natl. Cancer Inst. 1993;85:958–964. doi: 10.1093/jnci/85.12.958. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

