Distribution and Genomic Characterization of Third-Generation Cephalosporin-Resistant Escherichia coli Isolated from a Single Family and Home Environment: A 2-Year Longitudinal Study
- PMID: 36139932
- PMCID: PMC9495048
- DOI: 10.3390/antibiotics11091152
Distribution and Genomic Characterization of Third-Generation Cephalosporin-Resistant Escherichia coli Isolated from a Single Family and Home Environment: A 2-Year Longitudinal Study
Abstract
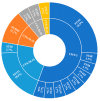
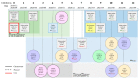
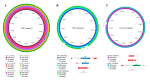
Third-generation cephalosporin-resistant Escherichia coli (CREC), particularly strains producing extended-spectrum β-lactamases (ESBLs), are a global concern. Our study aims to longitudinally assemble the genomic characteristics of CREC isolates from fecal samples from an index patient with recurrent CREC-related urinary tract infections and his family and swabs from his home environment 12 times between 2019 and 2021 to investigate the distribution of antibiotic resistance genes. CREC identified using the VITEK 2 were subjected to nanopore whole-genome sequencing (WGS). The WGS of 27 CREC isolates discovered in 137 specimens (1 urine, 123 feces, and 13 environmental) revealed the predominance of ST101 and ST131. Among these sequence types, blaCTX-M (44.4%, n = 12) was the predominant ESBL gene family, with blaCTX-M-14 (n = 6) being the most common. The remaining 15 (55.6%) isolates harbored blaCMY-2 genes and were clonally diverse. All E. coli isolated from the index patient's initial urine and fecal samples belonged to O25b:H4-B2-ST131 and carried blaCTX-M-14. The results of sequence analysis indicate plasmid-mediated household transmission of blaCMY-2 or blaCTX-M-55. A strong genomic similarity was discovered between fecal ESBL-producing E. coli and uropathogenic strains. Furthermore, blaCMY-2 genes were widely distributed among the CREC isolated from family members and their home environment.
Keywords: AmpC β-lactamases; CMY-2; Escherichia coli; extended-spectrum β-lactamases; plasmid; third-generation cephalosporin-resistant Escherichia coli; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders did not play any role in the study design, data collection and interpretation, or discussion to submit the study for publications.
Figures

References
-
- Imre K., Ban-Cucerzan A., Herman V., Sallam K.I., Cristina R.T., Abd-Elghany S.M., Morar D., Popa S.A., Imre M., Morar A. Occurrence, Pathogenic Potential and Antimicrobial Resistance of Escherichia coli Isolated from Raw Milk Cheese Commercialized in Banat Region, Romania. Antibiotics. 2022;11:721. doi: 10.3390/antibiotics11060721. - DOI - PMC - PubMed
-
- Smet A., Martel A., Persoons D., Dewulf J., Heyndrickx M., Herman L., Haesebrouck F., Butaye P. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: Molecular aspects, mobility and impact on public health. FEMS Microbiol. Rev. 2010;34:295–316. doi: 10.1111/j.1574-6976.2009.00198.x. - DOI - PubMed
-
- Sheng W.H., Badal R.E., Hsueh P.R. Distribution of extended-spectrum β-lactamases, AmpC β-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: Results of the study for Monitoring Antimicrobial Resistance Trends (SMART) Antimicrob. Agents Chemother. 2013;57:2981–2988. doi: 10.1128/aac.00971-12. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

