Adipocyte-Specific Laminin Alpha 4 Deletion Preserves Adipose Tissue Health despite Increasing Adiposity
- PMID: 36140178
- PMCID: PMC9495590
- DOI: 10.3390/biomedicines10092077
Adipocyte-Specific Laminin Alpha 4 Deletion Preserves Adipose Tissue Health despite Increasing Adiposity
Abstract
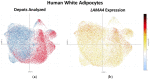
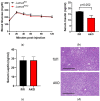
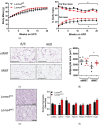
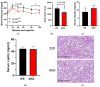
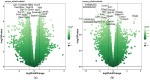
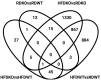
Laminins are heterotrimeric glycoproteins with structural and functional roles in basement membranes. The predominant laminin alpha chain found in adipocyte basement membranes is laminin α4 (LAMA4). Global LAMA4 deletion in mice leads to reduced adiposity and increased energy expenditure, but also results in vascular defects that complicate the interpretation of metabolic data. Here, we describe the generation and initial phenotypic analysis of an adipocyte-specific LAMA4 knockout mouse (Lama4AKO). We first performed an in-silico analysis to determine the degree to which laminin α4 was expressed in human and murine adipocytes. Next, male Lama4AKO and control mice were fed chow or high-fat diets and glucose tolerance was assessed along with serum insulin and leptin levels. Adipocyte area was measured in both epididymal and inguinal white adipose tissue (eWAT and iWAT, respectively), and eWAT was used for RNA-sequencing. We found that laminin α4 was highly expressed in human and murine adipocytes. Further, chow-fed Lama4AKO mice are like control mice in terms of body weight, body composition, and glucose tolerance, although they have larger eWAT adipocytes and lower insulin levels. High-fat-fed Lama4AKO mice are fatter and more glucose tolerant when compared to control mice. Transcriptionally, the eWAT of high-fat fed Lama4AKO mice resembles that of chow-fed control mice. We conclude from these findings that adipocyte-specific LAMA4 deletion is protective in an obesogenic environment, even though overall adiposity is increased.
Keywords: adipocyte; adipose tissue; basement membrane; extracellular matrix; laminin alpha 4; obesity.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures







Similar articles
-
Laminin α4 Expression in Human Adipose Tissue Depots and Its Association with Obesity and Obesity Related Traits.Biomedicines. 2023 Oct 17;11(10):2806. doi: 10.3390/biomedicines11102806. Biomedicines. 2023. PMID: 37893179 Free PMC article.
-
The Absence of Laminin α4 in Male Mice Results in Enhanced Energy Expenditure and Increased Beige Subcutaneous Adipose Tissue.Endocrinology. 2018 Jan 1;159(1):356-367. doi: 10.1210/en.2017-00186. Endocrinology. 2018. PMID: 28973559 Free PMC article.
-
Laminin-α4 Is Upregulated in Both Human and Murine Models of Obesity.Front Endocrinol (Lausanne). 2021 Jul 28;12:698621. doi: 10.3389/fendo.2021.698621. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34394003 Free PMC article.
-
Treatment with a SOD mimetic reduces visceral adiposity, adipocyte death, and adipose tissue inflammation in high fat-fed mice.Obesity (Silver Spring). 2014 Jan;22(1):178-87. doi: 10.1002/oby.20465. Epub 2013 Jun 13. Obesity (Silver Spring). 2014. PMID: 23526686 Free PMC article.
-
Laminin α4 deficient mice exhibit decreased capacity for adipose tissue expansion and weight gain.PLoS One. 2014 Oct 13;9(10):e109854. doi: 10.1371/journal.pone.0109854. eCollection 2014. PLoS One. 2014. PMID: 25310607 Free PMC article.
Cited by
-
The potential key genes within focal adhesion that regulate mesenchymal stem cells osteogenesis or adipogenesis in microgravity related disuse osteoporosis: an integrated analysis.Front Endocrinol (Lausanne). 2025 Mar 10;16:1469400. doi: 10.3389/fendo.2025.1469400. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40130165 Free PMC article.
-
Laminin α4 Expression in Human Adipose Tissue Depots and Its Association with Obesity and Obesity Related Traits.Biomedicines. 2023 Oct 17;11(10):2806. doi: 10.3390/biomedicines11102806. Biomedicines. 2023. PMID: 37893179 Free PMC article.
References
-
- Vaicik M.K., Blagajcevic A., Ye H., Morse M.C., Yang F., Goddi A., Brey E.M., Cohen R.N. The Absence of Laminin alpha4 in Male Mice Results in Enhanced Energy Expenditure and Increased Beige Subcutaneous Adipose Tissue. Endocrinology. 2018;159:356–367. doi: 10.1210/en.2017-00186. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

