Transgenic Expression of Nrf2 Induces a Pro-Reductive Stress and Adaptive Cardiac Remodeling in the Mouse
- PMID: 36140682
- PMCID: PMC9498410
- DOI: 10.3390/genes13091514
Transgenic Expression of Nrf2 Induces a Pro-Reductive Stress and Adaptive Cardiac Remodeling in the Mouse
Abstract
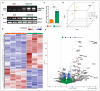
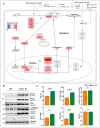
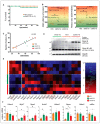
Nuclear factor, erythroid 2 like 2 (Nfe2l2 or Nrf2), is a transcription factor that protects cells by maintaining a homeostatic redox state during stress. The constitutive expression of Nrf2 (CaNrf2-TG) was previously shown to be pathological to the heart over time. We tested a hypothesis that the cardiac-specific expression of full length Nrf2 (mNrf2-TG) would moderately increase the basal antioxidant defense, triggering a pro-reductive environment leading to adaptive cardiac remodeling. Transgenic and non-transgenic (NTG) mice at 7−8 months of age were used to analyze the myocardial transcriptome, structure, and function. Next generation sequencing (NGS) for RNA profiling and qPCR-based validation of the NGS data, myocardial redox levels, and imaging (echocardiography) were performed. Transcriptomic analysis revealed that out of 14,665 identified mRNAs, 680 were differently expressed (DEG) in TG hearts. Of 680 DEGs, 429 were upregulated and 251 were downregulated significantly (FC > 2.0, p < 0.05). Gene set enrichment analysis revealed that the top altered pathways were (a) Nrf2 signaling, (b) glutathione metabolism and (c) ROS scavenging. A comparative analysis of the glutathione redox state in the hearts demonstrated significant differences between pro-reductive vs. hyper-reductive conditions (233 ± 36.7 and 380 ± 68.7 vs. 139 ± 8.6 µM/mg protein in mNrf2-TG and CaNrf2-TG vs. NTG). Genes involved in fetal development, hypertrophy, cytoskeletal rearrangement, histone deacetylases (HDACs), and GATA transcription factors were moderately increased in mNrf2-TG compared to CaNrf2-TG. Non-invasive echocardiography analysis revealed an increase in systolic function (ejection fraction) in mNrf2-TG, suggesting an adaptation, as opposed to pathological remodeling in CaNrf2-TG mice experiencing a hyper-reductive stress, leading to reduced survival (40% at 60 weeks). The effects of excess Nrf2-driven antioxidant transcriptome revealed a pro-reductive condition in the myocardium leading to an adaptive cardiac remodeling. While pre-conditioning the myocardial redox with excess antioxidants (i.e., pro-reductive state) could be beneficial against oxidative stress, a chronic pro-reductive environment in the myocardium might transition the adaptation to pathological remodeling.
Keywords: Nrf2 transgene; RNAseq; constitutively active Nrf2; echocardiography; reductive stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Nabeshima Y.I. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. - DOI - PubMed
-
- Kobayashi A., Kang M.-I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative Stress Sensor Keap1 Functions as an Adaptor for Cul3-Based E3 Ligase To Regulate Proteasomal Degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

