Next Generation Sequencing after Invasive Prenatal Testing in Fetuses with Congenital Malformations: Prenatal or Neonatal Investigation
- PMID: 36140685
- PMCID: PMC9498826
- DOI: 10.3390/genes13091517
Next Generation Sequencing after Invasive Prenatal Testing in Fetuses with Congenital Malformations: Prenatal or Neonatal Investigation
Abstract
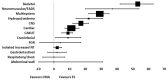
Congenital malformations diagnosed by ultrasound screening complicate 3-5% of pregnancies and many of these have an underlying genetic cause. Approximately 40% of prenatally diagnosed fetal malformations are associated with aneuploidy or copy number variants, detected by conventional karyotyping, QF-PCR and microarray techniques, however monogenic disorders are not diagnosed by these tests. Next generation sequencing as a secondary prenatal genetic test offers additional diagnostic yield for congenital abnormalities deemed to be potentially associated with an underlying genetic aetiology, as demonstrated by two large cohorts: the 'Prenatal assessment of genomes and exomes' (PAGE) study and 'Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study' performed at Columbia University in the US. These were large and prospective studies but relatively 'unselected' congenital malformations, with little Clinical Genetics input to the pre-test selection process. This review focuses on the incremental yield of next generation sequencing in single system congenital malformations, using evidence from the PAGE, Columbia and subsequent cohorts, with particularly high yields in those fetuses with cardiac and neurological anomalies, large nuchal translucency and non-immune fetal hydrops (of unknown aetiology). The total additional yield gained by exome sequencing in congenital heart disease was 12.7%, for neurological malformations 13.8%, 13.1% in increased nuchal translucency and 29% in non-immune fetal hydrops. This demonstrates significant incremental yield with exome sequencing in single-system anomalies and supports next generation sequencing as a secondary genetic test in routine clinical care of fetuses with congenital abnormalities.
Keywords: exome; fetus; genome; neonate; sequencing.
Conflict of interest statement
From 8 August 2022, MDK will be the Principal Senior Clinical Scientist for Illumina as well as remaining an Honorary Consultant in Fetal Medicine at Birmingham Women’s and Children’s Trust. The views expressed in this publication are those of the author and not necessarily those of Illumina. MDK is also a member of the RCOG Genomics Taskforce, the RCOG representative of the Joint Committee on Genomics in Medicine (joint committee of the Royal College of Physicians, Royal College of Pathologists, Royal College of Paediatrics and Child Health, Royal of Obstetricians and Gynaecologists) and a member of the Fetal Group of the British Society of Genetic Medicine.
Figures
References
-
- Mone F., McMullan D.J., Williams D., Chitty L.S., Maher E.R., Kilby M.D. Fetal Genomics Steering Group of the British Society for Genetic Medicine; on behalf of the Royal College of Obstetricians and Gynaecologists. Evidence to Support the Clinical Utility of Prenatal Exome Sequencing in Evaluation of the Fetus with Congenital Anomalies. Scientific Impact Paper No. 64. BJOG. 2021;128:e39–e50. doi: 10.1111/1471-0528.16616. - DOI - PubMed
-
- Lord J., McMullan D.J., Eberhardt R.Y., Rinck G., Hamilton S.J., Quinlan-Jones E., Prigmore E., Keelagher R., Best S.K., Carey G.K., et al. Prenatal Assessment of Genomes and Exomes Consortium. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): A cohort study. Lancet. 2019;393:747–757. doi: 10.1016/S0140-6736(18)31940-8. - DOI - PMC - PubMed
-
- Petrovski S., Aggarwal V., Giordano J.L., Stosic M., Wou K., Bier L., Spiegel E., Brennan K., Stong N., Jobanputra V., et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: A prospective cohort study. Lancet. 2019;393:758–767. doi: 10.1016/S0140-6736(18)32042-7. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources