Bacterial Communities Related to Aroma Formation during Spontaneous Fermentation of 'Cabernet Sauvignon' Wine in Ningxia, China
- PMID: 36140903
- PMCID: PMC9497756
- DOI: 10.3390/foods11182775
Bacterial Communities Related to Aroma Formation during Spontaneous Fermentation of 'Cabernet Sauvignon' Wine in Ningxia, China
Abstract
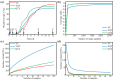
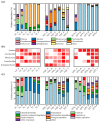
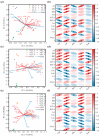
Bacteria are an important part of wine 'microbial terroir' and contribute to the formation of wine flavor. Based on high-throughput sequencing and non-targeted metabonomic technology, this study first explored the bacterial composition and its effect on the aroma formation of spontaneously fermented 'Cabernet Sauvignon' (CS) wine in the Eastern Foot of Helan Mountain (EFHM), Ningxia. The results showed that there were significant differences in bacterial communities during fermentation of CS grapes harvested from different sub-regions of EFHM, with the earlier-established vineyard obtaining more species. The level of bacterial diversity initially decreased and then increased as the fermentation proceeded. Malolactic fermentation (MLF) was spontaneously initiated during alcohol fermentation (AF). Pantoea, Lactobacillus, Rhodococcus, Fructobacillus, and Komagataeibacter were the core bacterial genera in the fermentation mixture. Lactobacillus contributed to the synthesis of methyl and isobutyl esters and the formation of red and black fruity fragrances of wine. Fructobacillus was closely related to the synthesis of aromatic alcohols and the generation of floral flavors.
Keywords: Cabernet Sauvignon; Ningxia; bacteria; flavor; spontaneous fermentation; wine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Co-evolutionary dynamics of microbial communities and flavor profiles during natural fermentation of Cabernet Sauvignon and Merlot: A comparative study within a single vineyard.Food Res Int. 2025 Jan;200:115517. doi: 10.1016/j.foodres.2024.115517. Epub 2024 Dec 15. Food Res Int. 2025. PMID: 39779148
-
Differences in Aroma Profile of Cabernet Sauvignon Grapes and Wines from Four Plots in Jieshi Mountain Region of Eastern China.Foods. 2023 Jul 11;12(14):2668. doi: 10.3390/foods12142668. Foods. 2023. PMID: 37509760 Free PMC article.
-
Influence of an indigenous yeast, CECA, from the Ningxia wine region of China, on the fungal and bacterial dynamics and function during Cabernet Sauvignon wine fermentation.J Sci Food Agric. 2024 Nov;104(14):8693-8706. doi: 10.1002/jsfa.13696. Epub 2024 Jun 26. J Sci Food Agric. 2024. PMID: 38922891
-
Wine flavor and aroma.J Ind Microbiol Biotechnol. 2011 Sep;38(9):1145-59. doi: 10.1007/s10295-011-1018-4. Epub 2011 Jul 24. J Ind Microbiol Biotechnol. 2011. PMID: 21786136 Review.
-
Aroma Precursors in Grapes and Wine: Flavor Release during Wine Production and Consumption.J Agric Food Chem. 2018 Mar 14;66(10):2281-2286. doi: 10.1021/acs.jafc.6b05255. Epub 2017 Mar 8. J Agric Food Chem. 2018. PMID: 28220693 Review.
Cited by
-
Comparison of microbial communities and volatile profiles of wines made from mulberry and grape.Appl Microbiol Biotechnol. 2023 Aug;107(16):5079-5094. doi: 10.1007/s00253-023-12632-y. Epub 2023 Jun 29. Appl Microbiol Biotechnol. 2023. PMID: 37382613
-
Application of Strain Selection Technology in Alcoholic Beverages: A Review.Foods. 2024 May 1;13(9):1396. doi: 10.3390/foods13091396. Foods. 2024. PMID: 38731767 Free PMC article. Review.
-
Volatile, Microbial, and Sensory Profiles and Consumer Acceptance of Coffee Cascara Kombuchas.Foods. 2023 Jul 15;12(14):2710. doi: 10.3390/foods12142710. Foods. 2023. PMID: 37509803 Free PMC article.
-
Characterization of the Flavors and Organoleptic Attributes of Petit Manseng Noble Rot Wines from the Eastern Foothills of Helan Mountain in Ningxia, China.Foods. 2025 Aug 4;14(15):2723. doi: 10.3390/foods14152723. Foods. 2025. PMID: 40807659 Free PMC article.
-
The bacterial succession and its role in flavor compounds formation during the fermentation of cigar tobacco leaves.Bioresour Bioprocess. 2023 Oct 31;10(1):74. doi: 10.1186/s40643-023-00694-9. Bioresour Bioprocess. 2023. PMID: 38647588 Free PMC article.
References
-
- Lappa I.K., Kachrimanidou V., Pateraki C., Koulougliotis D., Eriotou E., Kopsahelis N. Indigenous yeasts: Emerging trends and challenges in winemaking. Curr. Opin. Food Sci. 2020;32:133–143. doi: 10.1016/j.cofs.2020.04.004. - DOI
-
- Van Leeuwen C., Seguin G. The concept of terroir in viticulture. J. Wine Res. 2006;17:1–10. doi: 10.1080/09571260600633135. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

