A Combined Differential Proteome and Transcriptome Profiling of Fast- and Slow-Twitch Skeletal Muscle in Pigs
- PMID: 36140968
- PMCID: PMC9497725
- DOI: 10.3390/foods11182842
A Combined Differential Proteome and Transcriptome Profiling of Fast- and Slow-Twitch Skeletal Muscle in Pigs
Abstract
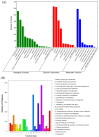
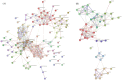
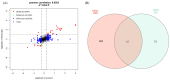
Skeletal muscle fiber types can contribute in part to affecting pork quality parameters. Biceps femoris (Bf) (fast muscle or white muscle) and Soleus (Sol) (slow muscle or red muscle) are two typical skeletal muscles characterized by obvious muscle fiber type differences in pigs. However, the critical proteins and potential regulatory mechanisms regulating porcine skeletal muscle fibers have yet to be clearly defined. In this study, the isobaric Tag for Relative and Absolute Quantification (iTRAQ)-based proteome was used to identify the key proteins affecting the skeletal muscle fiber types with Bf and Sol, by integrating the previous transcriptome data, while function enrichment analysis and a protein-protein interaction (PPI) network were utilized to explore the potential regulatory mechanisms of skeletal muscle fibers. A total of 126 differentially abundant proteins (DAPs) between the Bf and Sol were identified, and 12 genes were found to be overlapping between differentially expressed genes (DEGs) and DAPs, which are the critical proteins regulating the formation of skeletal muscle fibers. Functional enrichment and PPI analysis showed that the DAPs were mainly involved in the skeletal-muscle-associated structural proteins, mitochondria and energy metabolism, tricarboxylic acid cycle, fatty acid metabolism, and kinase activity, suggesting that PPI networks including DAPs are the main regulatory network affecting muscle fiber formation. Overall, these data provide valuable information for understanding the molecular mechanism underlying the formation and conversion of muscle fiber types, and provide potential markers for the evaluation of meat quality.
Keywords: meat quality; muscle fiber; pig; proteome; transcriptome.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Exploring the lncRNAs Related to Skeletal Muscle Fiber Types and Meat Quality Traits in Pigs.Genes (Basel). 2020 Aug 4;11(8):883. doi: 10.3390/genes11080883. Genes (Basel). 2020. PMID: 32759632 Free PMC article.
-
Phosphoproteomic analysis identifies differentially expressed phosphorylation sites that affect muscle fiber type in pigs.Front Nutr. 2022 Dec 22;9:1006739. doi: 10.3389/fnut.2022.1006739. eCollection 2022. Front Nutr. 2022. PMID: 36618708 Free PMC article.
-
Profiling and Functional Analysis of Circular RNAs in Porcine Fast and Slow Muscles.Front Cell Dev Biol. 2020 May 26;8:322. doi: 10.3389/fcell.2020.00322. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32528948 Free PMC article.
-
Muscle mechanics: adaptations with exercise-training.Exerc Sport Sci Rev. 1996;24:427-73. Exerc Sport Sci Rev. 1996. PMID: 8744258 Review.
-
Molecular mechanisms underlying the impact of muscle fiber types on meat quality in livestock and poultry.Front Vet Sci. 2023 Nov 22;10:1284551. doi: 10.3389/fvets.2023.1284551. eCollection 2023. Front Vet Sci. 2023. PMID: 38076559 Free PMC article. Review.
Cited by
-
Mass Spectrometry-Based Proteomic Technology and Its Application to Study Skeletal Muscle Cell Biology.Cells. 2023 Nov 1;12(21):2560. doi: 10.3390/cells12212560. Cells. 2023. PMID: 37947638 Free PMC article. Review.
-
Exploring Multi-Tissue Alternative Splicing and Skeletal Muscle Metabolism Regulation in Obese- and Lean-Type Pigs.Genes (Basel). 2024 Jan 31;15(2):196. doi: 10.3390/genes15020196. Genes (Basel). 2024. PMID: 38397185 Free PMC article.
-
Research Progress on the Regulating Factors of Muscle Fiber Heterogeneity in Livestock: A Review.Animals (Basel). 2024 Jul 31;14(15):2225. doi: 10.3390/ani14152225. Animals (Basel). 2024. PMID: 39123750 Free PMC article. Review.
References
-
- Su L., Li H., Xin X., Duan Y., Hua X., Jin Y. Muscle Fiber Types, Characteristics and Meat Quality. Adv. Mater. Res. 2013;634–638:1263–1267. doi: 10.4028/www.scientific.net/AMR.634-638.1263. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

