Multiple Perspectives Reveal the Role of DNA Damage Repair Genes in the Molecular Classification and Prognosis of Pancreatic Adenocarcinoma
- PMID: 36142142
- PMCID: PMC9499455
- DOI: 10.3390/ijms231810231
Multiple Perspectives Reveal the Role of DNA Damage Repair Genes in the Molecular Classification and Prognosis of Pancreatic Adenocarcinoma
Abstract
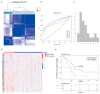
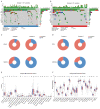
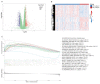
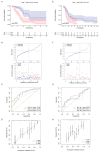
Pancreatic adenocarcinoma (PAAD) is a highly heterogeneous and immunosuppressive cancer. This study investigated the diversity of DNA damage repair (DDR) and immune microenvironment in PAAD by transcriptomic and genomic analysis. Patients with PAAD were divided into two DDR-based subtypes with distinct prognosis and molecular characteristics. The differential expression genes were mostly enriched in DDR and immune-related pathways. In order to distinguish high- and low-risk groups clinically, a DDR- and immune-based 5-gene prognostic signature (termed DPRS) was established. Patients in the high-risk group had inferior prognosis, a low level of immune checkpoint gene expression and low sensitivity to DDR-associated inhibitors. Furthermore, single-cell sequencing was used to observe the performance of the DDR-based signature in a high dimension, and immunohistochemistry was used to verify the relationship between the genes we identified and the prognosis of patients with PAAD. In conclusion, the DDR heterogeneity of PAAD was demonstrated, and a novel DDR- and immune-based risk-scoring model was constructed, which indicated the feasibility of DPRS in predicting prognosis and drug response in PAAD patients.
Keywords: DNA damage repair; immune; molecular classification; pancreatic adenocarcinoma; prognosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Development and Validation of an Inflammatory Response-Related Gene Signature for Predicting the Prognosis of Pancreatic Adenocarcinoma.Inflammation. 2022 Aug;45(4):1732-1751. doi: 10.1007/s10753-022-01657-6. Epub 2022 Mar 23. Inflammation. 2022. PMID: 35322324
-
Expression, potential biological behaviour and clinical significance of MCM3 in pancreatic adenocarcinoma: a comprehensive study integrating high throughput sequencing, CRISPR screening and in-house immunohistochemistry.Ann Med. 2024 Dec;56(1):2405879. doi: 10.1080/07853890.2024.2405879. Epub 2024 Sep 23. Ann Med. 2024. PMID: 39310930 Free PMC article.
-
Machine learning-based identification of biomarkers and drugs in immunologically cold and hot pancreatic adenocarcinomas.J Transl Med. 2024 Aug 16;22(1):775. doi: 10.1186/s12967-024-05590-0. J Transl Med. 2024. PMID: 39152432 Free PMC article.
-
Identify potential prognostic indicators and tumor-infiltrating immune cells in pancreatic adenocarcinoma.Biosci Rep. 2022 Feb 25;42(2):BSR20212523. doi: 10.1042/BSR20212523. Biosci Rep. 2022. PMID: 35083488 Free PMC article. Review.
-
The molecular biology of pancreatic adenocarcinoma: translational challenges and clinical perspectives.Signal Transduct Target Ther. 2021 Jul 5;6(1):249. doi: 10.1038/s41392-021-00659-4. Signal Transduct Target Ther. 2021. PMID: 34219130 Free PMC article. Review.
Cited by
-
Metabolic reprogramming and prognostic modeling in pancreatic cancer: insights from WGCNA.Front Genet. 2025 Jun 12;16:1487046. doi: 10.3389/fgene.2025.1487046. eCollection 2025. Front Genet. 2025. PMID: 40574800 Free PMC article.
References
-
- Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G.H., Hoadley K.A., Rashid N.U., Williams L.A., Eaton S.C., Chung A.H., et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

