Gut Microbiota Functional Traits, Blood pH, and Anti-GAD Antibodies Concur in the Clinical Characterization of T1D at Onset
- PMID: 36142163
- PMCID: PMC9499637
- DOI: 10.3390/ijms231810256
Gut Microbiota Functional Traits, Blood pH, and Anti-GAD Antibodies Concur in the Clinical Characterization of T1D at Onset
Abstract
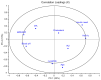
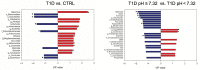
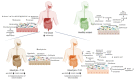
Alterations of gut microbiota have been identified before clinical manifestation of type 1 diabetes (T1D). To identify the associations amongst gut microbiome profile, metabolism and disease markers, the 16S rRNA-based microbiota profiling and 1H-NMR metabolomic analysis were performed on stool samples of 52 T1D patients at onset, 17 T1D siblings and 57 healthy subjects (CTRL). Univariate, multivariate analyses and classification models were applied to clinical and -omic integrated datasets. In T1D patients and their siblings, Clostridiales and Dorea were increased and Dialister and Akkermansia were decreased compared to CTRL, while in T1D, Lachnospiraceae were higher and Collinsella was lower, compared to siblings and CTRL. Higher levels of isobutyrate, malonate, Clostridium, Enterobacteriaceae, Clostridiales, Bacteroidales, were associated to T1D compared to CTRL. Patients with higher anti-GAD levels showed low abundances of Roseburia, Faecalibacterium and Alistipes and those with normal blood pH and low serum HbA1c levels showed high levels of purine and pyrimidine intermediates. We detected specific gut microbiota profiles linked to both T1D at the onset and to diabetes familiarity. The presence of specific microbial and metabolic profiles in gut linked to anti-GAD levels and to blood acidosis can be considered as predictive biomarker associated progression and severity of T1D.
Keywords: anti-GAD antibody; gut microbiota ecology and metabolome; insulin need; ketoacidosis; microbial biomarkers; omics data integration; pediatrics; type 1 diabetes (T1D).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

