Simultaneous Probing of Metabolism and Oxygenation of Tumors In Vivo Using FLIM of NAD(P)H and PLIM of a New Polymeric Ir(III) Oxygen Sensor
- PMID: 36142177
- PMCID: PMC9499414
- DOI: 10.3390/ijms231810263
Simultaneous Probing of Metabolism and Oxygenation of Tumors In Vivo Using FLIM of NAD(P)H and PLIM of a New Polymeric Ir(III) Oxygen Sensor
Abstract
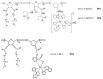
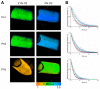
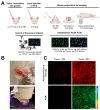
Tumor cells are well adapted to grow in conditions of variable oxygen supply and hypoxia by switching between different metabolic pathways. However, the regulatory effect of oxygen on metabolism and its contribution to the metabolic heterogeneity of tumors have not been fully explored. In this study, we develop a methodology for the simultaneous analysis of cellular metabolic status, using the fluorescence lifetime imaging microscopy (FLIM) of metabolic cofactor NAD(P)H, and oxygen level, using the phosphorescence lifetime imaging (PLIM) of a new polymeric Ir(III)-based sensor (PIr3) in tumors in vivo. The sensor, derived from a polynorbornene and cyclometalated iridium(III) complex, exhibits the oxygen-dependent quenching of phosphorescence with a 40% longer lifetime in degassed compared to aerated solutions. In vitro, hypoxia resulted in a correlative increase in PIr3 phosphorescence lifetime and free (glycolytic) NAD(P)H fraction in cells. In vivo, mouse tumors demonstrated a high degree of cellular-level heterogeneity of both metabolic and oxygen states, and a lower dependence of metabolism on oxygen than cells in vitro. The small tumors were hypoxic, while the advanced tumors contained areas of normoxia and hypoxia, which was consistent with the pimonidazole assay and angiographic imaging. Dual FLIM/PLIM metabolic/oxygen imaging will be valuable in preclinical investigations into the effects of hypoxia on metabolic aspects of tumor progression and treatment response.
Keywords: bioimaging; in vitro; in vivo; oxygen sensing; phosphorescence lifetime imaging; phosphorescent polymeric iridium(III) complexes; tumor.
Conflict of interest statement
V.I.S. was affiliated with Becker & Hickl GmbH, Germany, when conducting the research.
Figures









References
-
- Vaupel P., Flood A.B., Swartz H.M. Oxygenation Status of Malignant Tumors vs. Normal Tissues: Critical Evaluation and Updated Data Source Based on Direct Measurements with pO2 Microsensors. Appl. Magn. Reson. 2021;52:1451–1479. doi: 10.1007/s00723-021-01383-6. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

