Progesterone Receptor Modulates Extraembryonic Mesoderm and Cardiac Progenitor Specification during Mouse Gastrulation
- PMID: 36142249
- PMCID: PMC9499561
- DOI: 10.3390/ijms231810307
Progesterone Receptor Modulates Extraembryonic Mesoderm and Cardiac Progenitor Specification during Mouse Gastrulation
Abstract
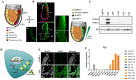
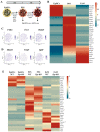
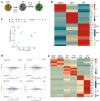
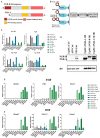
Progesterone treatment is commonly employed to promote and support pregnancy. While maternal tissues are the main progesterone targets in humans and mice, its receptor (PGR) is expressed in the murine embryo, questioning its function during embryonic development. Progesterone has been previously associated with murine blastocyst development. Whether it contributes to lineage specification is largely unknown. Gastrulation initiates lineage specification and generation of the progenitors contributing to all organs. Cells passing through the primitive streak (PS) will give rise to the mesoderm and endoderm. Cells emerging posteriorly will form the extraembryonic mesodermal tissues supporting embryonic growth. Cells arising anteriorly will contribute to the embryonic heart in two sets of distinct progenitors, first (FHF) and second heart field (SHF). We found that PGR is expressed in a posterior-anterior gradient in the PS of gastrulating embryos. We established in vitro differentiation systems inducing posterior (extraembryonic) and anterior (cardiac) mesoderm to unravel PGR function. We discovered that PGR specifically modulates extraembryonic and cardiac mesoderm. Overexpression experiments revealed that PGR safeguards cardiac differentiation, blocking premature SHF progenitor specification and sustaining the FHF progenitor pool. This role of PGR in heart development indicates that progesterone administration should be closely monitored in potential early-pregnancy patients undergoing infertility treatment.
Keywords: ECM; cardiac differentiation; cell adhesion; epiblast stem cells; extraembryonic mesoderm; mesoderm induction; mouse gastrulation; progesterone receptor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Sim C.B., Phipson B., Ziemann M., Rafehi H., Mills R.J., Watt K.I., Abu-Bonsrah K.D., Kalathur R.K., Voges H.K., Dinh D.T., et al. Sex-Specific Control of Human Heart Maturation by the Progesterone Receptor. Circulation. 2021;143:1614–1628. doi: 10.1161/CIRCULATIONAHA.120.051921. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

