Crystal Structure of an Archaeal Tyrosyl-tRNA Synthetase Bound to Photocaged L-Tyrosine and Its Potential Application to Time-Resolved X-ray Crystallography
- PMID: 36142308
- PMCID: PMC9499402
- DOI: 10.3390/ijms231810399
Crystal Structure of an Archaeal Tyrosyl-tRNA Synthetase Bound to Photocaged L-Tyrosine and Its Potential Application to Time-Resolved X-ray Crystallography
Abstract
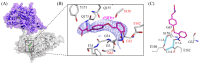
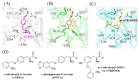
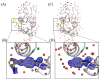
Genetically encoded caged amino acids can be used to control the dynamics of protein activities and cellular localization in response to external cues. In the present study, we revealed the structural basis for the recognition of O-(2-nitrobenzyl)-L-tyrosine (oNBTyr) by its specific variant of Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (oNBTyrRS), and then demonstrated its potential availability for time-resolved X-ray crystallography. The substrate-bound crystal structure of oNBTyrRS at a 2.79 Å resolution indicated that the replacement of tyrosine and leucine at positions 32 and 65 by glycine (Tyr32Gly and Leu65Gly, respectively) and Asp158Ser created sufficient space for entry of the bulky substitute into the amino acid binding pocket, while Glu in place of Leu162 formed a hydrogen bond with the nitro moiety of oNBTyr. We also produced an oNBTyr-containing lysozyme through a cell-free protein synthesis system derived from the Escherichia coli B95. ΔA strain with the UAG codon reassigned to the nonnatural amino acid. Another crystallographic study of the caged protein showed that the site-specifically incorporated oNBTyr was degraded to tyrosine by light irradiation of the crystals. Thus, cell-free protein synthesis of caged proteins with oNBTyr could facilitate time-resolved structural analysis of proteins, including medically important membrane proteins.
Keywords: O-(2-nitrobenzyl)-L-tyrosine; X-ray crystallography; caged amino acid; cell-free protein synthesis; nonnatural amino acid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

