Ustilaginoidea virens Nuclear Effector SCRE4 Suppresses Rice Immunity via Inhibiting Expression of a Positive Immune Regulator OsARF17
- PMID: 36142440
- PMCID: PMC9501289
- DOI: 10.3390/ijms231810527
Ustilaginoidea virens Nuclear Effector SCRE4 Suppresses Rice Immunity via Inhibiting Expression of a Positive Immune Regulator OsARF17
Abstract
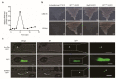
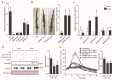
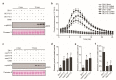
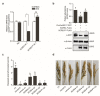
Rice false smut caused by the biotrophic fungal pathogen Ustilaginoidea virens has become one of the most important diseases in rice. The large effector repertory in U. virens plays a crucial role in virulence. However, current knowledge of molecular mechanisms how U. virens effectors target rice immune signaling to promote infection is very limited. In this study, we identified and characterized an essential virulence effector, SCRE4 (Secreted Cysteine-Rich Effector 4), in U. virens. SCRE4 was confirmed as a secreted nuclear effector through yeast secretion, translocation assays and protein subcellular localization, as well as up-regulation during infection. The SCRE4 gene deletion attenuated the virulence of U. virens to rice. Consistently, ectopic expression of SCRE4 in rice inhibited chitin-triggered immunity and enhanced susceptibility to false smut, substantiating that SCRE4 is an essential virulence factor. Furthermore, SCRE4 transcriptionally suppressed the expression of OsARF17, an auxin response factor in rice, which positively regulates rice immune responses and resistance against U. virens. Additionally, the immunosuppressive capacity of SCRE4 depended on its nuclear localization. Therefore, we uncovered a virulence strategy in U. virens that transcriptionally suppresses the expression of the immune positive modulator OsARF17 through nucleus-localized effector SCRE4 to facilitate infection.
Keywords: Ustilaginoidea virens; auxin response factor 17; rice false smut; secreted cysteine-rich effector 4; transcription inhibition.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Tanaka E., Ashizawa T., Sonoda R., Tanaka C. Villosiclava virens gen. nov., comb. nov., teleomorph of Ustilaginoidea virens, the causal agent of rice false smut. Mycotaxon. 2008;106:491–501.
-
- Tang Y.X., Jin J., Hu D.W., Yong M.L., Xu Y., He L.P. Elucidation of the infection process of Ustilaginoidea virens (teleomorph: Villosiclava virens) in rice spikelets. Plant Pathol. 2013;62:1–8. doi: 10.1111/j.1365-3059.2012.02629.x. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

