Comprehensive Transcriptomic and Proteomic Analyses Identify a Candidate Gene Set in Cross-Resistance for Endocrine Therapy in Breast Cancer
- PMID: 36142451
- PMCID: PMC9501051
- DOI: 10.3390/ijms231810539
Comprehensive Transcriptomic and Proteomic Analyses Identify a Candidate Gene Set in Cross-Resistance for Endocrine Therapy in Breast Cancer
Abstract
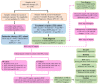
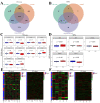
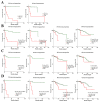
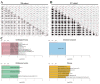
Endocrine therapy (ET) of selective estrogen receptor modulators (SERMs), selective estrogen receptor downregulators (SERDs), and aromatase inhibitors (AIs) has been used as the gold standard treatment for hormone-receptor-positive (HR+) breast cancer. Despite its clinical benefits, approximately 30% of patients develop ET resistance, which remains a major clinical challenge in patients with HR+ breast cancer. The mechanisms of ET resistance mainly focus on mutations in the ER and related pathways; however, other targets still exist from ligand-independent ER reactivation. Moreover, mutations in the ER that confer resistance to SERMs or AIs seldom appear in SERDs. To date, little research has been conducted to identify a critical target that appears in both SERMs/SERDs and AIs. In this study, we conducted comprehensive transcriptomic and proteomic analyses from two cohorts of The Cancer Genome Atlas Breast Invasive Carcinoma (TCGA-BRCA) to identify the critical targets for both SERMs/SERDs and AIs of ET resistance. From a treatment response cohort with treatment response for the initial ET regimen and an endocrine therapy cohort with survival outcomes, we identified candidate gene sets that appeared in both SERMs/SERDs and AIs of ET resistance. The candidate gene sets successfully differentiated progress/resistant groups (PD) from complete response groups (CR) and were significantly correlated with survival outcomes in both cohorts. In summary, this study provides valuable clinical implications for the critical roles played by candidate gene sets in the diagnosis, mechanism, and therapeutic strategy for both SERMs/SERDs and AIs of ET resistance for the future.
Keywords: The Cancer Genome Atlas (TCGA); aromatase inhibitors (AIs); breast cancer; cross-resistance; endocrine therapy resistance; selective estrogen receptor degraders (SERDs); selective estrogen receptor modulators (SERMs).
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the study design; collection, analyses, or interpretation of data; writing of the manuscript, or decision to publish the results.
Figures

Similar articles
-
The race to develop oral SERDs and other novel estrogen receptor inhibitors: recent clinical trial results and impact on treatment options.Cancer Metastasis Rev. 2022 Dec;41(4):975-990. doi: 10.1007/s10555-022-10066-y. Epub 2022 Oct 14. Cancer Metastasis Rev. 2022. PMID: 36229710 Free PMC article. Review.
-
Accelerating drug development in breast cancer: New frontiers for ER inhibition.Cancer Treat Rev. 2022 Sep;109:102432. doi: 10.1016/j.ctrv.2022.102432. Epub 2022 Jun 27. Cancer Treat Rev. 2022. PMID: 35839531 Review.
-
Autophagy and senescence facilitate the development of antiestrogen resistance in ER positive breast cancer.Front Endocrinol (Lausanne). 2024 Mar 18;15:1298423. doi: 10.3389/fendo.2024.1298423. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38567308 Free PMC article. Review.
-
Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment.Pharmacol Ther. 2018 Jun;186:1-24. doi: 10.1016/j.pharmthera.2017.12.012. Epub 2017 Dec 28. Pharmacol Ther. 2018. PMID: 29289555 Review.
-
Selective Estrogen Receptor Degraders (SERDs): A Promising Strategy for Estrogen Receptor Positive Endocrine-Resistant Breast Cancer.J Med Chem. 2020 Dec 24;63(24):15094-15114. doi: 10.1021/acs.jmedchem.0c00913. Epub 2020 Nov 2. J Med Chem. 2020. PMID: 33138369
Cited by
-
Multiomics insights on the onset, progression, and metastatic evolution of breast cancer.Front Oncol. 2023 Dec 19;13:1292046. doi: 10.3389/fonc.2023.1292046. eCollection 2023. Front Oncol. 2023. PMID: 38169859 Free PMC article. Review.
-
Molecular Research and Treatment of Breast Cancer 2.0.Int J Mol Sci. 2024 Apr 1;25(7):3932. doi: 10.3390/ijms25073932. Int J Mol Sci. 2024. PMID: 38612742 Free PMC article.
References
-
- Hammond M.E., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S., Fitzgibbons P.L., Francis G., Goldstein N.S., Hayes M., et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

