Vesicular Glutamate Release from Feeder-FreehiPSC-Derived Neurons
- PMID: 36142455
- PMCID: PMC9501332
- DOI: 10.3390/ijms231810545
Vesicular Glutamate Release from Feeder-FreehiPSC-Derived Neurons
Abstract
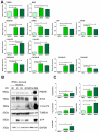
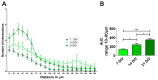
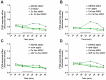
Human-induced pluripotent stem cells (hiPSCs) represent one of the main and powerful tools for the in vitro modeling of neurological diseases. Standard hiPSC-based protocols make use of animal-derived feeder systems to better support the neuronal differentiation process. Despite their efficiency, such protocols may not be appropriate to dissect neuronal specific properties or to avoid interspecies contaminations, hindering their future translation into clinical and drug discovery approaches. In this work, we focused on the optimization of a reproducible protocol in feeder-free conditions able to generate functional glutamatergic neurons. This protocol is based on a generation of neuroprecursor cells differentiated into human neurons with the administration in the culture medium of specific neurotrophins in a Geltrex-coated substrate. We confirmed the efficiency of this protocol through molecular analysis (upregulation of neuronal markers and neurotransmitter receptors assessed by gene expression profiling and expression of the neuronal markers at the protein level), morphological analysis, and immunfluorescence detection of pre-synaptic and post-synaptic markers at synaptic boutons. The hiPSC-derived neurons acquired Ca2+-dependent glutamate release properties as a hallmark of neuronal maturation. In conclusion, our study describes a new methodological approach to achieve feeder-free neuronal differentiation from hiPSC and adds a new tool for functional characterization of hiPSC-derived neurons.
Keywords: diseases modelling; human neurons; human-induced pluripotent stem cells (hiPSC); neurotransmitter release; stem cells.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures




References
-
- Krewski D., Acosta D., Jr., Andersen M., Anderson H., Bailar J.C., 3rd, Boekelheide K., Brent R., Charnley G., Cheung V.G., Green S., Jr., et al. Toxicity testing in the 21st century: A vision and a strategy. J. Toxicol. Environ. Health B Crit. Rev. 2010;13:51–138. doi: 10.1080/10937404.2010.483176. - DOI - PMC - PubMed
-
- Consiglio E.D., Pistollato F., Gyves E.M.-D., Bal-Price A., Testai E. Integrating biokinetics and in vitro studies to evaluate developmental neurotoxicity induced by chlorpyrifos in human iPSC-derived neural stem cells undergoing differentiation towards neuronal and glial cells. Reprod. Toxicol. 2020;98:174–188. doi: 10.1016/j.reprotox.2020.09.010. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- Cycle 2019b - Project #1916 to F.Z./The Jerome Lejeune Foundation
- Ricerca Corrente to F.Z./Italian Ministry of Health
- FFABR (Fondo di finanziamento per le attività base di ricerca) to C.C./MIUR-Italy
- FFABR (Fondo di finanziamento per le attività base di ricerca) to M.M./MIUR-Italy
- Ph.D School of the Department of Experimental Medicine contribution 2021 to S.A./University of Genova
LinkOut - more resources
Full Text Sources
Miscellaneous

