Identification of the Regulatory Targets of miR-3687 and miR-4417 in Prostate Cancer Cells Using a Proteomics Approach
- PMID: 36142477
- PMCID: PMC9501364
- DOI: 10.3390/ijms231810565
Identification of the Regulatory Targets of miR-3687 and miR-4417 in Prostate Cancer Cells Using a Proteomics Approach
Abstract
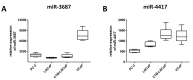
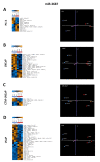
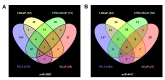
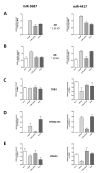
MicroRNAs (miRNA) are ubiquitous non-coding RNAs that have a prominent role in cellular regulation. The expression of many miRNAs is often found deregulated in prostate cancer (PCa) and castration-resistant prostate cancer (CRPC). Although their expression can be associated with PCa and CRPC, their functions and regulatory activity in cancer development are poorly understood. In this study, we used different proteomics tools to analyze the activity of hsa-miR-3687-3p (miR-3687) and hsa-miR-4417-3p (miR-4417), two miRNAs upregulated in CRPC. PCa and CRPC cell lines were transfected with miR-3687 or miR-4417 to overexpress the miRNAs. Cell lysates were analyzed using 2D gel electrophoresis and proteins were subsequently identified using mass spectrometry (Maldi-MS/MS). A whole cell lysate, without 2D-gel separation, was analyzed by ESI-MS/MS. The expression of deregulated proteins found across both methods was further investigated using Western blotting. Gene ontology and cellular process network analysis determined that miR-3687 and miR-4417 are involved in diverse regulatory mechanisms that support the CRPC phenotype, including metabolism and inflammation. Moreover, both miRNAs are associated with extracellular vesicles, which point toward a secretory mechanism. The tumor protein D52 isoform 1 (TD52-IF1), which regulates neuroendocrine trans-differentiation, was found to be substantially deregulated in androgen-insensitive cells by both miR-3687 and miR-4417. These findings show that these miRNAs potentially support the CRPC by truncating the TD52-IF1 expression after the onset of androgen resistance.
Keywords: castration-resistant prostate cancer; cellular regulation; microRNA; prostate cancer; proteomics.
Conflict of interest statement
The authors declare no conflict of interest regarding the present study.
Figures





Similar articles
-
Upregulation of miR-3195, miR-3687 and miR-4417 is associated with castration-resistant prostate cancer.World J Urol. 2021 Oct;39(10):3789-3797. doi: 10.1007/s00345-021-03723-4. Epub 2021 May 14. World J Urol. 2021. PMID: 33990872 Free PMC article.
-
Functional roles of miR-625-5p and miR-874-3p in the progression of castration resistant prostate cancer.Life Sci. 2022 Jul 15;301:120603. doi: 10.1016/j.lfs.2022.120603. Epub 2022 May 1. Life Sci. 2022. PMID: 35508255
-
Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer.Br J Cancer. 2017 Jul 25;117(3):409-420. doi: 10.1038/bjc.2017.191. Epub 2017 Jun 22. Br J Cancer. 2017. PMID: 28641312 Free PMC article.
-
The Crosstalk of Long Non-Coding RNA and MicroRNA in Castration-Resistant and Neuroendocrine Prostate Cancer: Their Interaction and Clinical Importance.Int J Mol Sci. 2021 Dec 30;23(1):392. doi: 10.3390/ijms23010392. Int J Mol Sci. 2021. PMID: 35008817 Free PMC article. Review.
-
MicroRNA Regulation of Androgen Receptor in Castration-Resistant Prostate Cancer: Premises, Promises, and Potentials.Curr Mol Pharmacol. 2021 Oct 25;14(4):559-569. doi: 10.2174/1874467213666201223121850. Curr Mol Pharmacol. 2021. PMID: 33357209 Review.
Cited by
-
Bioinformatics Analysis of the Prognostic Significance of CAND1 in ERα-Positive Breast Cancer.Diagnostics (Basel). 2022 Sep 27;12(10):2327. doi: 10.3390/diagnostics12102327. Diagnostics (Basel). 2022. PMID: 36292029 Free PMC article.
-
Microfluidic-based prostate cancer model for investigating the secretion of prostate-specific antigen and microRNAs in vitro.Sci Rep. 2023 Jul 19;13(1):11623. doi: 10.1038/s41598-023-38834-y. Sci Rep. 2023. PMID: 37468746 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

