Transcriptional Activation of Ecdysone-Responsive Genes Requires H3K27 Acetylation at Enhancers
- PMID: 36142704
- PMCID: PMC9502983
- DOI: 10.3390/ijms231810791
Transcriptional Activation of Ecdysone-Responsive Genes Requires H3K27 Acetylation at Enhancers
Abstract
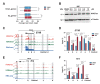
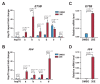
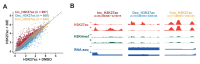
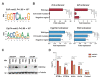
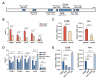
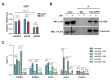
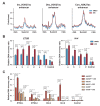
The steroid hormone ecdysone regulates insect development via its nuclear receptor (the EcR protein), which functions as a ligand-dependent transcription factor. The EcR regulates target gene expression by binding to ecdysone response elements (EcREs) in their promoter or enhancer regions. Its role in epigenetic regulation and, particularly, in histone acetylation remains to be clarified. Here, we analyzed the dynamics of histone acetylation and demonstrated that the acetylation of histone H3 on lysine 27 (H3K27) at enhancers was required for the transcriptional activation of ecdysone-responsive genes. Western blotting and ChIP-qPCR revealed that ecdysone altered the acetylation of H3K27. For E75B and Hr4, ecdysone-responsive genes, enhancer activity, and transcription required the histone acetyltransferase activity of the CBP. EcR binding was critical in inducing enhancer activity and H3K27 acetylation. The CREB-binding protein (CBP) HAT domain catalyzed H3K27 acetylation and CBP coactivation with EcR, independent of the presence of ecdysone. Increased H3K27 acetylation promoted chromatin accessibility, with the EcR and CBP mediating a local chromatin opening in response to ecdysone. Hence, epigenetic mechanisms, including the modification of acetylation and chromatin accessibility, controlled ecdysone-dependent gene transcription.
Keywords: chromatin regulation; cisregulatory elements; ecdysone receptor; gene transcription; histone modification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Ecdysone induced gene expression is associated with acetylation of histone H3 lysine 23 in Drosophila melanogaster.PLoS One. 2012;7(7):e40565. doi: 10.1371/journal.pone.0040565. Epub 2012 Jul 10. PLoS One. 2012. PMID: 22808194 Free PMC article.
-
CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing.Development. 2009 Sep;136(18):3131-41. doi: 10.1242/dev.037127. Development. 2009. PMID: 19700617 Free PMC article.
-
Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters.Genome Res. 2012 Jun;22(6):1081-8. doi: 10.1101/gr.136929.111. Epub 2012 Apr 16. Genome Res. 2012. PMID: 22508764 Free PMC article.
-
Genomic occupancy of the transcriptional co-activators p300 and CBP.Transcription. 2013 Jan-Feb;4(1):18-23. doi: 10.4161/trns.22601. Epub 2012 Nov 6. Transcription. 2013. PMID: 23131664 Free PMC article. Review.
-
The chromatin signatures of enhancers and their dynamic regulation.Nucleus. 2023 Dec;14(1):2160551. doi: 10.1080/19491034.2022.2160551. Nucleus. 2023. PMID: 36602897 Free PMC article. Review.
Cited by
-
The Drosophila Zinc Finger Protein Aef1 Colocalizes with Enhancers and Is Involved in the Transcriptional Regulation of Numerous Genes.Acta Naturae. 2025 Apr-Jun;17(2):58-63. doi: 10.32607/actanaturae.27556. Acta Naturae. 2025. PMID: 40771843 Free PMC article.
-
Coregulators Reside within Drosophila Ecdysone-Inducible Loci before and after Ecdysone Treatment.Int J Mol Sci. 2023 Jul 24;24(14):11844. doi: 10.3390/ijms241411844. Int J Mol Sci. 2023. PMID: 37511602 Free PMC article.
-
Changes in gene expression levels caused by H3K9me3/H3K9ac modifications are associated with BmCPV infection in Bombyx mori.Virulence. 2025 Dec;16(1):2510535. doi: 10.1080/21505594.2025.2510535. Epub 2025 May 26. Virulence. 2025. PMID: 40418637 Free PMC article.
-
Multifaceted regulatory mechanisms of the EGR family in tumours and prospects for therapeutic applications (Review).Int J Mol Med. 2025 Jul;56(1):113. doi: 10.3892/ijmm.2025.5554. Epub 2025 May 30. Int J Mol Med. 2025. PMID: 40444475 Free PMC article. Review.
-
SGK1 aggravates idiopathic pulmonary fibrosis by triggering H3k27ac-mediated macrophage reprogramming and disturbing immune homeostasis.Int J Biol Sci. 2024 Jan 12;20(3):968-986. doi: 10.7150/ijbs.90808. eCollection 2024. Int J Biol Sci. 2024. PMID: 38250161 Free PMC article.
References
-
- Nishita Y. Ecdysone response elements in the distal promoter of the Bombyx Broad-Complex gene, BmBR-C. Insect Mol. Biol. 2014;23:341–356. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

