OsHsfB4b Confers Enhanced Drought Tolerance in Transgenic Arabidopsis and Rice
- PMID: 36142741
- PMCID: PMC9501395
- DOI: 10.3390/ijms231810830
OsHsfB4b Confers Enhanced Drought Tolerance in Transgenic Arabidopsis and Rice
Abstract
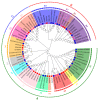
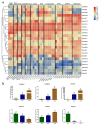
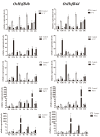
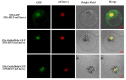
Heat shock factors (Hsfs) play pivotal roles in plant stress responses and confer stress tolerance. However, the functions of several Hsfs in rice (Oryza sativa L.) are not yet known. In this study, genome-wide analysis of the Hsf gene family in rice was performed. A total of 25 OsHsf genes were identified, which could be clearly clustered into three major groups, A, B, and C, based on the characteristics of the sequences. Bioinformatics analysis showed that tandem duplication and fragment replication were two important driving forces in the process of evolution and expansion of the OsHsf family genes. Both OsHsfB4b and OsHsfB4d showed strong responses to the stress treatment. The results of subcellular localization showed that the OsHsfB4b protein was in the nucleus whereas the OsHsfB4d protein was located in both the nucleus and cytoplasm. Over-expression of the OsHsfB4b gene in Arabidopsis and rice can increase the resistance to drought stress. This study provides a basis for understanding the function and evolutionary history of the OsHsf gene family, enriching our knowledge of understanding the biological functions of OsHsfB4b and OsHsfB4d genes involved in the stress response in rice, and also reveals the potential value of OsHsfB4b in rice environmental adaptation improvement.
Keywords: Hsf; drought stress; expression profiles; genome-wide analysis; rice.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses.BMC Genomics. 2014 Nov 21;15(1):1009. doi: 10.1186/1471-2164-15-1009. BMC Genomics. 2014. PMID: 25416131 Free PMC article.
-
A systematic view of rice heat shock transcription factor family using phylogenomic analysis.J Plant Physiol. 2013 Feb 15;170(3):321-9. doi: 10.1016/j.jplph.2012.09.008. Epub 2012 Oct 31. J Plant Physiol. 2013. PMID: 23122336
-
Rice OsHSFA3 Gene Improves Drought Tolerance by Modulating Polyamine Biosynthesis Depending on Abscisic Acid and ROS Levels.Int J Mol Sci. 2020 Mar 9;21(5):1857. doi: 10.3390/ijms21051857. Int J Mol Sci. 2020. PMID: 32182761 Free PMC article.
-
OsHsfA2c and OsHsfB4b are involved in the transcriptional regulation of cytoplasmic OsClpB (Hsp100) gene in rice (Oryza sativa L.).Cell Stress Chaperones. 2012 Mar;17(2):243-54. doi: 10.1007/s12192-011-0303-5. Epub 2011 Nov 1. Cell Stress Chaperones. 2012. PMID: 22147560 Free PMC article.
-
[Biological characteristics of heat shock transcription factors and their roles in abiotic stress adaptation of higher plant].Ying Yong Sheng Tai Xue Bao. 2022 Aug;33(8):2286-2296. doi: 10.13287/i.1001-9332.202208.039. Ying Yong Sheng Tai Xue Bao. 2022. PMID: 36043838 Review. Chinese.
Cited by
-
Comprehensive co-expression network reveals the fine-tuning of AsHSFA2c in balancing drought tolerance and growth in oat.Commun Biol. 2025 Mar 8;8(1):393. doi: 10.1038/s42003-025-07857-8. Commun Biol. 2025. PMID: 40057657 Free PMC article.
-
Characterization of the Heat Shock Transcription Factor Family in Lycoris radiata and Its Potential Roles in Response to Abiotic Stresses.Plants (Basel). 2024 Jan 17;13(2):271. doi: 10.3390/plants13020271. Plants (Basel). 2024. PMID: 38256823 Free PMC article.
-
Genome-Wide Analysis of the Hsf Gene Family in Rosa chinensis and RcHsf17 Function in Thermotolerance.Int J Mol Sci. 2024 Dec 31;26(1):287. doi: 10.3390/ijms26010287. Int J Mol Sci. 2024. PMID: 39796142 Free PMC article.
-
Biocuration of a Transcription Factors Network Involved in Submergence Tolerance during Seed Germination and Coleoptile Elongation in Rice (Oryza sativa).Plants (Basel). 2023 May 29;12(11):2146. doi: 10.3390/plants12112146. Plants (Basel). 2023. PMID: 37299125 Free PMC article.
-
Characterization of the heat shock factor RcHsfA6 in Rosa chinensis and function in the thermotolerance of Arabidopsis.BMC Plant Biol. 2025 May 21;25(1):673. doi: 10.1186/s12870-025-06652-1. BMC Plant Biol. 2025. PMID: 40399821 Free PMC article.
References
-
- Zhou R., Li B., Liu H., Sun D. Progress in the participation of Ca2+–calmodulin in heat shock signal transduction. Prog. Nat. Sci. 2009;19:1201–1208. doi: 10.1016/j.pnsc.2008.12.011. - DOI
MeSH terms
Substances
Grants and funding
- No. 30601733/High-level talents of Henan Agricultural University
- No. 2021DG700024-KF202218/State Key Laboratory for Managing Biotic and Chemical Treats to the Quality and Safety of Agro-products
- No: 2021DG700024-KF202218/State Key Laboratory of Agricultural Product Quality and Safety Hazard Factors and Risk Prevention and Control
LinkOut - more resources
Full Text Sources

