A Single Aspergillus fumigatus Gene Enables Ergothioneine Biosynthesis and Secretion by Saccharomyces cerevisiae
- PMID: 36142753
- PMCID: PMC9502471
- DOI: 10.3390/ijms231810832
A Single Aspergillus fumigatus Gene Enables Ergothioneine Biosynthesis and Secretion by Saccharomyces cerevisiae
Abstract
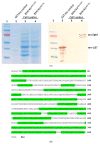
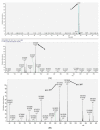
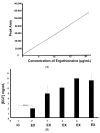
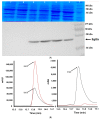
The naturally occurring sulphur-containing histidine derivative, ergothioneine (EGT), exhibits potent antioxidant properties and has been proposed to confer human health benefits. Although it is only produced by select fungi and prokaryotes, likely to protect against environmental stress, the GRAS organism Saccharomyces cerevisiae does not produce EGT naturally. Herein, it is demonstrated that the recombinant expression of a single gene, Aspergillus fumigatus egtA, in S. cerevisiae results in EgtA protein presence which unexpectedly confers complete EGT biosynthetic capacity. Both High Performance Liquid Chromatography (HPLC) and LC−mass spectrometry (MS) analysis were deployed to detect and confirm EGT production in S. cerevisiae. The localisation and quantification of the resultant EGT revealed a significantly (p < 0.0001) larger quantity of EGT was extracellularly present in culture supernatants than intracellularly accumulated in 96 h yeast cultures. Methionine addition to cultures improved EGT production. The additional expression of two candidate cysteine desulfurases from A. fumigatus was thought to be required to complete EGT biosynthesis, namely AFUA_2G13295 and AFUA_3G14240, termed egt2a and egt2b in this study. However, the co-expression of egtA and egt2a in S. cerevisiae resulted in a significant decrease in the observed EGT levels (p < 0.05). The AlphaFold prediction of A. fumigatus EgtA 3-Dimensional structure illuminates the bidomain structure of the enzyme and the opposing locations of both active sites. Overall, we clearly show that recombinant S. cerevisiae can biosynthesise and secrete EGT in an EgtA-dependent manner which presents a facile means of producing EGT for biotechnological and biomedical use.
Keywords: AlphaFold; ROS; antioxidant; cell factory; ergothioneine; redox stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Biosynthesis of ergothioneine: current state, achievements, and perspectives.Appl Microbiol Biotechnol. 2025 Apr 12;109(1):93. doi: 10.1007/s00253-025-13476-4. Appl Microbiol Biotechnol. 2025. PMID: 40220171 Free PMC article. Review.
-
Ergothioneine Biosynthesis and Functionality in the Opportunistic Fungal Pathogen, Aspergillus fumigatus.Sci Rep. 2016 Oct 17;6:35306. doi: 10.1038/srep35306. Sci Rep. 2016. PMID: 27748436 Free PMC article.
-
Successful biosynthesis of natural antioxidant ergothioneine in Saccharomyces cerevisiae required only two genes from Grifola frondosa.Microb Cell Fact. 2020 Aug 18;19(1):164. doi: 10.1186/s12934-020-01421-1. Microb Cell Fact. 2020. PMID: 32811496 Free PMC article.
-
Ergothioneine production using Methylobacterium species, yeast, and fungi.J Biosci Bioeng. 2018 Dec;126(6):715-722. doi: 10.1016/j.jbiosc.2018.05.021. Epub 2018 Jun 14. J Biosci Bioeng. 2018. PMID: 29910189
-
Involvement of Sulfur in the Biosynthesis of Essential Metabolites in Pathogenic Fungi of Animals, Particularly Aspergillus spp.: Molecular and Therapeutic Implications.Front Microbiol. 2019 Dec 13;10:2859. doi: 10.3389/fmicb.2019.02859. eCollection 2019. Front Microbiol. 2019. PMID: 31921039 Free PMC article. Review.
Cited by
-
Linker-mediated domain separation enhances cold adaptation in cellulases.Protein Sci. 2025 Aug;34(8):e70198. doi: 10.1002/pro.70198. Protein Sci. 2025. PMID: 40671324 Free PMC article.
-
Biosynthesis of ergothioneine: current state, achievements, and perspectives.Appl Microbiol Biotechnol. 2025 Apr 12;109(1):93. doi: 10.1007/s00253-025-13476-4. Appl Microbiol Biotechnol. 2025. PMID: 40220171 Free PMC article. Review.
-
CRISPR/Cas9-based iterative multi-copy integration for improved metabolite yields in Saccharomyces cerevisiae.Synth Syst Biotechnol. 2025 Mar 5;10(2):629-637. doi: 10.1016/j.synbio.2025.02.016. eCollection 2025 Jun. Synth Syst Biotechnol. 2025. PMID: 40151793 Free PMC article.
References
-
- Bello M.H., Barrera-Perez V., Morin D., Epstein L. The Neurospora crassa mutant NcDeltaEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet. Biol. 2012;49:160–172. doi: 10.1016/j.fgb.2011.12.007. - DOI - PubMed
-
- Gamage A.M., Liao C., Cheah I.K., Chen Y., Lim D.R.X., Ku J.W.K., Chee R.S.L., Gengenbacher M., Seebeck F.P., Halliwell B., et al. The proteobacterial species Burkholderia pseudomallei produces ergothioneine, which enhances virulence in mammalian infection. FASEB J. 2018;32:6395–6409. doi: 10.1096/fj.201800716. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

