A Comprehensive Evolutionary Study of Chloroplast RNA Editing in Gymnosperms: A Novel Type of G-to-A RNA Editing Is Common in Gymnosperms
- PMID: 36142757
- PMCID: PMC9505161
- DOI: 10.3390/ijms231810844
A Comprehensive Evolutionary Study of Chloroplast RNA Editing in Gymnosperms: A Novel Type of G-to-A RNA Editing Is Common in Gymnosperms
Abstract
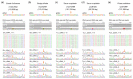
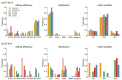
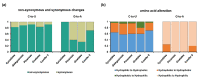
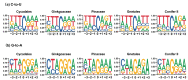
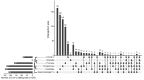
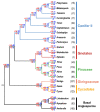
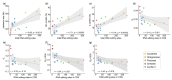
Although more than 9100 plant plastomes have been sequenced, RNA editing sites of the whole plastome have been experimentally verified in only approximately 21 species, which seriously hampers the comprehensive evolutionary study of chloroplast RNA editing. We investigated the evolutionary pattern of chloroplast RNA editing sites in 19 species from all 13 families of gymnosperms based on a combination of genomic and transcriptomic data. We found that the chloroplast C-to-U RNA editing sites of gymnosperms shared many common characteristics with those of other land plants, but also exhibited many unique characteristics. In contrast to that noted in angiosperms, the density of RNA editing sites in ndh genes was not the highest in the sampled gymnosperms, and both loss and gain events at editing sites occurred frequently during the evolution of gymnosperms. In addition, GC content and plastomic size were positively correlated with the number of chloroplast RNA editing sites in gymnosperms, suggesting that the increase in GC content could provide more materials for RNA editing and facilitate the evolution of RNA editing in land plants or vice versa. Interestingly, novel G-to-A RNA editing events were commonly found in all sampled gymnosperm species, and G-to-A RNA editing exhibits many different characteristics from C-to-U RNA editing in gymnosperms. This study revealed a comprehensive evolutionary scenario for chloroplast RNA editing sites in gymnosperms, and reported that a novel type of G-to-A RNA editing is prevalent in gymnosperms.
Keywords: G-to-A; GC content; RNA editing; chloroplast; gymnosperms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata.RNA. 2014 Oct;20(10):1499-506. doi: 10.1261/rna.045575.114. Epub 2014 Aug 20. RNA. 2014. PMID: 25142065 Free PMC article.
-
Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: the pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms.Proc Natl Acad Sci U S A. 1996 Aug 6;93(16):8766-70. doi: 10.1073/pnas.93.16.8766. Proc Natl Acad Sci U S A. 1996. PMID: 8710946 Free PMC article.
-
Chloroplast C-to-U RNA editing in vascular plants is adaptive due to its restorative effect: testing the restorative hypothesis.RNA. 2023 Feb;29(2):141-152. doi: 10.1261/rna.079450.122. RNA. 2023. PMID: 36649983 Free PMC article.
-
[RNA editing in chloroplasts: a trail of the genome evolution?].Tanpakushitsu Kakusan Koso. 2005 Nov;50(14 Suppl):1858-9. Tanpakushitsu Kakusan Koso. 2005. PMID: 16318339 Review. Japanese. No abstract available.
-
Plastid ndh genes in plant evolution.Plant Physiol Biochem. 2010 Aug;48(8):636-45. doi: 10.1016/j.plaphy.2010.04.009. Epub 2010 May 9. Plant Physiol Biochem. 2010. PMID: 20493721 Review.
Cited by
-
The Taihangia mitogenome provides new insights into its adaptation and organelle genome evolution in Rosaceae.Planta. 2025 Feb 12;261(3):59. doi: 10.1007/s00425-025-04629-w. Planta. 2025. PMID: 39939538
-
Piece and parcel of gymnosperm organellar genomes.Planta. 2024 Jun 3;260(1):14. doi: 10.1007/s00425-024-04449-4. Planta. 2024. PMID: 38829418 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

