Comparative Genomic Analysis of Agarolytic Flavobacterium faecale WV33T
- PMID: 36142798
- PMCID: PMC9501601
- DOI: 10.3390/ijms231810884
Comparative Genomic Analysis of Agarolytic Flavobacterium faecale WV33T
Abstract
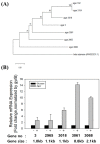
Flavobacteria are widely dispersed in a variety of environments and produce various polysaccharide-degrading enzymes. Here, we report the complete genome of Flavobacterium faecale WV33T, an agar-degrading bacterium isolated from the stools of Antarctic penguins. The sequenced genome of F. faecale WV33T represents a single circular chromosome (4,621,116 bp, 35.2% G + C content), containing 3984 coding DNA sequences and 85 RNA-coding genes. The genome of F. faecale WV33T contains 154 genes that encode carbohydrate-active enzymes (CAZymes). Among the CAZymes, seven putative genes encoding agarases have been identified in the genome. Transcriptional analysis revealed that the expression of these putative agarases was significantly enhanced by the presence of agar in the culture medium, suggesting that these proteins are involved in agar hydrolysis. Pangenome analysis revealed that the genomes of the 27 Flavobacterium type strains, including F. faecale WV33T, tend to be very plastic, and Flavobacterium strains are unique species with a tiny core genome and a large non-core region. The average nucleotide identity and phylogenomic analysis of the 27 Flavobacterium-type strains showed that F. faecale WV33T was positioned in a unique clade in the evolutionary tree.
Keywords: CAZymes; Flavobacterium faecale; agarase; comparative genomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Flavobacterium faecale sp. nov., an agarase-producing species isolated from stools of Antarctic penguins.Int J Syst Evol Microbiol. 2014 Aug;64(Pt 8):2884-2890. doi: 10.1099/ijs.0.059618-0. Epub 2014 Jun 3. Int J Syst Evol Microbiol. 2014. PMID: 24893942
-
A Novel Auxiliary Agarolytic Pathway Expands Metabolic Versatility in the Agar-Degrading Marine Bacterium Colwellia echini A3T.Appl Environ Microbiol. 2021 May 26;87(12):e0023021. doi: 10.1128/AEM.00230-21. Epub 2021 May 26. Appl Environ Microbiol. 2021. PMID: 33811026 Free PMC article.
-
Flavobacterium litorale sp. nov., isolated from red alga.Int J Syst Evol Microbiol. 2022 Aug;72(8). doi: 10.1099/ijsem.0.005458. Int J Syst Evol Microbiol. 2022. PMID: 35951659
-
Genomic insights and comparative analysis of Flavobacterium bizetiae HJ-32-4 isolated from soil.Antonie Van Leeuwenhoek. 2023 Oct;116(10):975-986. doi: 10.1007/s10482-023-01858-5. Epub 2023 Aug 5. Antonie Van Leeuwenhoek. 2023. PMID: 37542623
-
Flavobacterium ginsenosidimutans sp. nov., a bacterium with ginsenoside converting activity isolated from soil of a ginseng field.Int J Syst Evol Microbiol. 2011 Jun;61(Pt 6):1408-1412. doi: 10.1099/ijs.0.025700-0. Epub 2010 Jul 2. Int J Syst Evol Microbiol. 2011. PMID: 20601491
Cited by
-
Pangenome analysis provides insights into the genetic diversity, metabolic versatility, and evolution of the genus Flavobacterium.Microbiol Spectr. 2023 Aug 18;11(5):e0100323. doi: 10.1128/spectrum.01003-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37594286 Free PMC article.
References
-
- Bernardet J.F., Bowman J.P., Genus I. Flavobacterium. In: Krieg N.R., Ludwig W., Whitman W., Hedlund B.P., Paster B.J., Staley J.T., Ward N., Brown D., Parte A., editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Volume 4. Springer; New York, NY, USA: 2011. pp. 112–154.
-
- Bernardet J.F., Nakagawa Y. An introduction to the family Flavobacteriaceae. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K., Stackebrandt E., editors. The Prokaryotes, a Handbook on the Biology of Bacteria. 3rd ed. Volume 7. Springer; New York, NY, USA: 2006. pp. 455–480.
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

