Characterization of a mGluR5 Knockout Rat Model with Hallmarks of Fragile X Syndrome
- PMID: 36143345
- PMCID: PMC9504063
- DOI: 10.3390/life12091308
Characterization of a mGluR5 Knockout Rat Model with Hallmarks of Fragile X Syndrome
Abstract
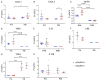
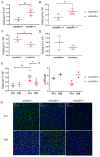
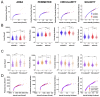
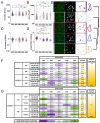
The number of reported cases of neurodevelopmental disorders has increased significantly in the last few decades, but the etiology of these diseases remains poorly understood. There is evidence of a fundamental link between genetic abnormalities and symptoms of autism spectrum disorders (ASDs), and the most common monogenetic inheritable form of ASDs is Fragile X Syndrome (FXS). Previous studies indicate that FXS is linked to glutamate signaling regulation by the G-protein-coupled metabotropic glutamate receptor 5 (mGluR5), which has been shown to have a regulatory role in neuroinflammation. We characterized the effect of knocking out mGluR5 in an organism known to have complex cognitive functions-the rat. The heterozygous phenotype is the most clinically relevant; therefore, we performed analysis in heterozygous pups. We showed developmental abnormalities in heterozygous mGluR5 knockout rats, as well as a significant increase in chemokine (C-X-C motif) ligand 1 (CXCL) expression, a hallmark indicator of early onset inflammation. We quantified an increase in microglial density in the knockout pups and quantified morphological phenotypes representative of greater reactivity in the male vs. female and postnatal day 28 heterozygous pups compared to postnatal day 14 heterozygous pups. In response to injury, reactive microglia release matrix metalloproteases, contribute to extracellular matrix (ECM) breakdown, and are responsible for eradicating cellular and molecular debris. In our study, the changes in microglial density and reactivity correlated with abnormalities in the mRNA expression levels of ECM proteins and with the density of perineuronal nets. We saw atypical neuropsychiatric behavior in open field and elevated plus tests in heterozygous pups compared to wild-type litter and age-matched controls. These results demonstrate the pathological potential of the mGluR5 knockout in rats and further support the presence of neuroinflammatory roots in ASDs.
Keywords: autism spectrum disorders (ASDs); extracellular matrix; machine learning; microglia; neurodevelopmental disorders; neuroinflammation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Decreased expression of mGluR5 within the dorsolateral prefrontal cortex in autism and increased microglial number in mGluR5 knockout mice: Pathophysiological and neurobehavioral implications.Brain Behav Immun. 2015 Oct;49:197-205. doi: 10.1016/j.bbi.2015.05.009. Epub 2015 Jun 5. Brain Behav Immun. 2015. PMID: 26052099
-
Is metabotropic glutamate receptor 5 upregulated in prefrontal cortex in fragile X syndrome?Mol Autism. 2013 May 24;4(1):15. doi: 10.1186/2040-2392-4-15. Mol Autism. 2013. PMID: 23706040 Free PMC article.
-
Selective Disruption of Metabotropic Glutamate Receptor 5-Homer Interactions Mimics Phenotypes of Fragile X Syndrome in Mice.J Neurosci. 2016 Feb 17;36(7):2131-47. doi: 10.1523/JNEUROSCI.2921-15.2016. J Neurosci. 2016. PMID: 26888925 Free PMC article.
-
Convergent evidence for mGluR5 in synaptic and neuroinflammatory pathways implicated in ASD.Neurosci Biobehav Rev. 2015 May;52:172-7. doi: 10.1016/j.neubiorev.2015.02.006. Epub 2015 Feb 19. Neurosci Biobehav Rev. 2015. PMID: 25704074 Review.
-
Role for metabotropic glutamate receptor 5 (mGluR5) in the pathogenesis of fragile X syndrome.J Physiol. 2008 Mar 15;586(6):1503-8. doi: 10.1113/jphysiol.2008.150722. Epub 2008 Jan 17. J Physiol. 2008. PMID: 18202092 Free PMC article. Review.
Cited by
-
Metabotropic glutamate receptors (mGluRs) in epileptogenesis: an update on abnormal mGluRs signaling and its therapeutic implications.Neural Regen Res. 2024 Feb;19(2):360-368. doi: 10.4103/1673-5374.379018. Neural Regen Res. 2024. PMID: 37488891 Free PMC article. Review.
References
-
- ASD Data and Statistics. [(accessed on 21 July 2022)];2021 Available online: https://www.cdc.gov/ncbddd/autism/data/index.html.
-
- Autism and Developmental Disabilities Monitoring Network Surveillance Year 2008 Principal Investigators Prevalence of Autism Spectrum Disorders—Autism and Developmental Disabilities Monitoring Network, 14 Sites, United States, 2008. Surveill. Summ. 2012;61:19. - PubMed
-
- Kado Y., Sanada S., Oono S., Ogino T., Nouno S. Children with autism spectrum disorder comorbid with attention-deficit/hyperactivity disorder examined by the Wisconsin card sorting test: Analysis by age-related differences. Brain Dev. 2020;42:113–120. doi: 10.1016/j.braindev.2019.07.011. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

