SARS-CoV-2 and HIV: Impact on Pulmonary Epithelial Cells
- PMID: 36143354
- PMCID: PMC9500782
- DOI: 10.3390/life12091317
SARS-CoV-2 and HIV: Impact on Pulmonary Epithelial Cells
Abstract
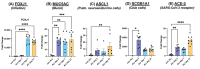
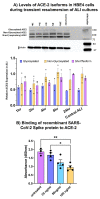
The SARS-CoV-2 pandemic provides a natural opportunity for the collision of coronavirus disease-2019 (COVID-19) with chronic infections, which place numerous individuals at high risk of severe COVID-19. Infection with Human Immunodeficiency Virus (HIV), a global epidemic, remains a major public health concern. Whether prior HIV+ status exacerbates COVID-19 warrants investigation. Herein, we characterized the impact of SARS-CoV-2 in human bronchial epithelial cells (HBECs) previously exposed to HIV. We optimized the air-liquid interface (ALI) cell culture technique to allow for challenges with HIV at the basolateral cell surface and SARS-CoV-2 spike protein on the apical surface, followed by genetic analyses for cellular stress/toxicity and innate/adaptive immune responses. Our results suggest that the IL-10 pathway was consistently activated in HBECs treated with spike, HIV, or a combination. Recombinant spike protein elicited COVID-19 cytokine storms while HIV activated different signaling pathways. HIV-treated HBECs could no longer activate NF-kB, pro-inflammatory TRAF-6 ubiquitination nor RIP1 signaling. Combinations of HIV and SARS-CoV-2 spike increased gene expression for activation of endoplasmic reticulum-phagosome pathway and downregulated non-canonical NF-kB pathways that are key in functional regulatory T cells and RNA Polymerase II transcription. Our in vitro studies suggest that prior HIV infection may not exacerbate COVID-19. Further in vivo studies are warranted to advance this field.
Keywords: HIV; SARS-CoV-2; adaptive; air liquid interphase (ALI); co-infection; immune; innate; polyparasitism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Differential Effect of SARS-CoV-2 Spike Glycoprotein 1 on Human Bronchial and Alveolar Lung Mucosa Models: Implications for Pathogenicity.Viruses. 2021 Dec 17;13(12):2537. doi: 10.3390/v13122537. Viruses. 2021. PMID: 34960806 Free PMC article.
-
Human surfactant protein D facilitates SARS-CoV-2 pseudotype binding and entry in DC-SIGN expressing cells, and downregulates spike protein induced inflammation.Front Immunol. 2022 Jul 28;13:960733. doi: 10.3389/fimmu.2022.960733. eCollection 2022. Front Immunol. 2022. PMID: 35967323 Free PMC article.
-
Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein finds additional vaccine-induced epitopes beyond those for mild infection.Elife. 2022 Jan 24;11:e73490. doi: 10.7554/eLife.73490. Elife. 2022. PMID: 35072628 Free PMC article.
-
COVID-19 and HIV-Associated Immune Reconstitution Inflammatory Syndrome: Emergence of Pathogen-Specific Immune Responses Adding Fuel to the Fire.Front Immunol. 2021 Mar 24;12:649567. doi: 10.3389/fimmu.2021.649567. eCollection 2021. Front Immunol. 2021. PMID: 33841434 Free PMC article. Review.
-
Insights into COVID-19 Vaccine Development Based on Immunogenic Structural Proteins of SARS-CoV-2, Host Immune Responses, and Herd Immunity.Cells. 2021 Oct 29;10(11):2949. doi: 10.3390/cells10112949. Cells. 2021. PMID: 34831172 Free PMC article. Review.
Cited by
-
SARS-CoV-2 spike treatment and transfection impairs airway epithelial repair.ERJ Open Res. 2025 Aug 4;11(4):00940-2024. doi: 10.1183/23120541.00940-2024. eCollection 2025 Jul. ERJ Open Res. 2025. PMID: 40761650 Free PMC article.
References
-
-
UNAIDS. UNAIDS Data 2021. 2022.
-
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

