Transcriptome Analysis Provides Insights into Potentilla bifurca Adaptation to High Altitude
- PMID: 36143374
- PMCID: PMC9503701
- DOI: 10.3390/life12091337
Transcriptome Analysis Provides Insights into Potentilla bifurca Adaptation to High Altitude
Abstract
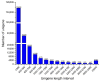
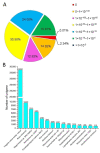
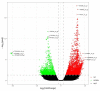
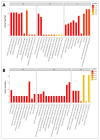
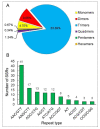
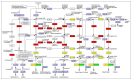
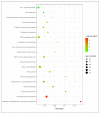
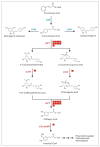
Potentilla bifurca is widely distributed in Eurasia, including the Tibetan Plateau. It is a valuable medicinal plant in the Tibetan traditional medicine system, especially for the treatment of diabetes. This study investigated the functional gene profile of Potentilla bifurca at different altitudes by RNA-sequencing technology, including de novo assembly of 222,619 unigenes from 405 million clean reads, 57.64% of which were annotated in Nr, GO, KEGG, Pfam, and Swiss-Prot databases. The most significantly differentially expressed top 50 genes in the high-altitude samples were derived from plants that responded to abiotic stress, such as peroxidase, superoxide dismutase protein, and the ubiquitin-conjugating enzyme. Pathway analysis revealed that a large number of DEGs encode key enzymes involved in secondary metabolites, including phenylpropane and flavonoids. In addition, a total of 298 potential genomic SSRs were identified in this study, which provides information on the development of functional molecular markers for genetic diversity assessment. In conclusion, this study provides the first comprehensive assessment of the Potentilla bifurca transcriptome. This provides new insights into coping mechanisms for non-model organisms surviving in harsh environments at high altitudes, as well as molecular evidence for the selection of superior medicinal plants.
Keywords: Potentilla bifurca; de novo transcriptome sequencing; flavonoid metabolism; high altitude; secondary metabolism.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
De novo transcriptome characterization of the ghost moth, Thitarodes pui, and elevation-based differences in the gene expression of its larvae.Gene. 2015 Dec 10;574(1):95-105. doi: 10.1016/j.gene.2015.07.084. Epub 2015 Jul 31. Gene. 2015. PMID: 26235680
-
Physiological, biochemical and proteomics analysis reveals the adaptation strategies of the alpine plant Potentilla saundersiana at altitude gradient of the Northwestern Tibetan Plateau.J Proteomics. 2015 Jan 1;112:63-82. doi: 10.1016/j.jprot.2014.08.009. Epub 2014 Aug 30. J Proteomics. 2015. PMID: 25181701
-
Comparative transcriptome analysis of aboveground and underground tissues of Rhodiola algida, an important ethno-medicinal herb endemic to the Qinghai-Tibetan Plateau.Gene. 2014 Dec 15;553(2):90-7. doi: 10.1016/j.gene.2014.09.063. Gene. 2014. PMID: 25281820
-
High-throughput transcriptome sequencing analysis provides preliminary insights into the biotransformation mechanism of Rhodopseudomonas palustris treated with alpha-rhamnetin-3-rhamnoside.Microbiol Res. 2016 Apr;185:1-12. doi: 10.1016/j.micres.2016.01.002. Epub 2016 Jan 22. Microbiol Res. 2016. PMID: 26946373
-
Comparative transcriptome among Euscaphis konishii Hayata tissues and analysis of genes involved in flavonoid biosynthesis and accumulation.BMC Genomics. 2019 Jan 9;20(1):24. doi: 10.1186/s12864-018-5354-x. BMC Genomics. 2019. PMID: 30626333 Free PMC article.
Cited by
-
Adaptation of High-Altitude Plants to Harsh Environments: Application of Phenotypic-Variation-Related Methods and Multi-Omics Techniques.Int J Mol Sci. 2024 Nov 26;25(23):12666. doi: 10.3390/ijms252312666. Int J Mol Sci. 2024. PMID: 39684378 Free PMC article. Review.
-
Transcriptional landscape illustrates the diversified adaptation of medicinal plants to multifactorial stress combinations linked with high altitude.Planta. 2025 Apr 15;261(5):111. doi: 10.1007/s00425-025-04686-1. Planta. 2025. PMID: 40234266
-
Insights into the Adaptation to High Altitudes from Transcriptome Profiling: A Case Study of an Endangered Species, Kingdonia uniflora.Genes (Basel). 2023 Jun 19;14(6):1291. doi: 10.3390/genes14061291. Genes (Basel). 2023. PMID: 37372473 Free PMC article.
-
Comparative metabolomics of two nettle species unveils distinct high-altitude adaptation mechanisms on the Tibetan Plateau.BMC Plant Biol. 2025 May 15;25(1):640. doi: 10.1186/s12870-025-06666-9. BMC Plant Biol. 2025. PMID: 40375155 Free PMC article.
-
Metabolites of Geum aleppicum and Sibbaldianthe bifurca: Diversity and α-Glucosidase Inhibitory Potential.Metabolites. 2023 May 25;13(6):689. doi: 10.3390/metabo13060689. Metabolites. 2023. PMID: 37367847 Free PMC article.
References
-
- Magana U.R., Escudero A., Gavilan R.G. Metabolic and physiological responses of Mediterranean high-mountain and alpine plants to combined abiotic stresses. Physiol. Plant. 2019;165:403–412. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

