Discovery of Potential SARS-CoV-2 Papain-like Protease Natural Inhibitors Employing a Multi-Phase In Silico Approach
- PMID: 36143445
- PMCID: PMC9505301
- DOI: 10.3390/life12091407
Discovery of Potential SARS-CoV-2 Papain-like Protease Natural Inhibitors Employing a Multi-Phase In Silico Approach
Abstract
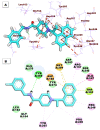
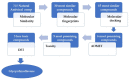
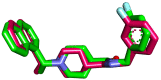
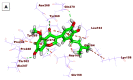
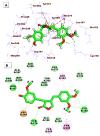
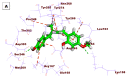
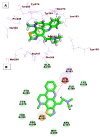
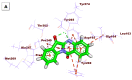
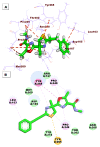
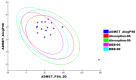
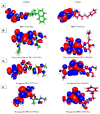
As an extension of our research against COVID-19, a multiphase in silico approach was applied in the selection of the three most common inhibitors (Glycyrrhizoflavone (76), Arctigenin (94), and Thiangazole (298)) against papain-like protease, PLpro (PDB ID: 4OW0), among 310 metabolites of natural origin. All compounds of the exam set were reported as antivirals. The structural similarity between the examined compound set and S88, the co-crystallized ligand of PLpro, was examined through structural similarity and fingerprint studies. The two experiments pointed to Brevicollin (28), Cryptopleurine (41), Columbamine (46), Palmatine (47), Glycyrrhizoflavone (76), Licochalcone A (87), Arctigenin (94), Termilignan (98), Anolignan B (99), 4,5-dihydroxy-6″-deoxybromotopsentin (192), Dercitin (193), Tryptanthrin (200), 6-Cyano-5-methoxy-12-methylindolo [2, 3A] carbazole (211), Thiangazole (298), and Phenoxan (300). The binding ability against PLpro was screened through molecular docking, disclosing the favorable binding modes of six metabolites. ADMET studies expected molecules 28, 76, 94, 200, and 298 as the most favorable metabolites. Then, molecules 76, 94, and 298 were chosen through in silico toxicity studies. Finally, DFT studies were carried out on glycyrrhizoflavone (76) and indicated a high level of similarity in the molecular orbital analysis. The obtained data can be used in further in vitro and in vivo studies to examine and confirm the inhibitory effect of the filtered metabolites against PLpro and SARS-CoV-2.
Keywords: ADMET; DFT; SARS-CoV-2; molecular docking; natural products; papain-like protease; structural similarity.
Conflict of interest statement
No conflict of interest to be declared.
Figures








Similar articles
-
The Discovery of Potential SARS-CoV-2 Natural Inhibitors among 4924 African Metabolites Targeting the Papain-like Protease: A Multi-Phase In Silico Approach.Metabolites. 2022 Nov 16;12(11):1122. doi: 10.3390/metabo12111122. Metabolites. 2022. PMID: 36422263 Free PMC article.
-
In Silico Screening of Semi-Synthesized Compounds as Potential Inhibitors for SARS-CoV-2 Papain-like Protease: Pharmacophoric Features, Molecular Docking, ADMET, Toxicity and DFT Studies.Molecules. 2021 Oct 30;26(21):6593. doi: 10.3390/molecules26216593. Molecules. 2021. PMID: 34771004 Free PMC article.
-
Microbial based natural compounds as potential inhibitors for SARS-CoV-2 Papain-like protease (PLpro): a molecular docking and dynamic simulation study.J Biomol Struct Dyn. 2022;40(24):13848-13858. doi: 10.1080/07391102.2021.1997815. Epub 2021 Nov 3. J Biomol Struct Dyn. 2022. PMID: 34730069
-
Chalcone-amide, a privileged backbone for the design and development of selective SARS-CoV/SARS-CoV-2 papain-like protease inhibitors.Eur J Med Chem. 2022 Oct 5;240:114572. doi: 10.1016/j.ejmech.2022.114572. Epub 2022 Jul 3. Eur J Med Chem. 2022. PMID: 35797899 Free PMC article. Review.
-
Potential Papain-like Protease Inhibitors Against COVID-19: A Comprehensive In Silico Based Review.Comb Chem High Throughput Screen. 2022;25(11):1838-1858. doi: 10.2174/1386207325666211122123602. Comb Chem High Throughput Screen. 2022. PMID: 34809541 Review.
Cited by
-
The Discovery of Potential SARS-CoV-2 Natural Inhibitors among 4924 African Metabolites Targeting the Papain-like Protease: A Multi-Phase In Silico Approach.Metabolites. 2022 Nov 16;12(11):1122. doi: 10.3390/metabo12111122. Metabolites. 2022. PMID: 36422263 Free PMC article.
-
The Essential Oil of Petroselinum crispum (Mill) Fuss Seeds from Peru: Phytotoxic Activity and In Silico Evaluation on the Target Enzyme of the Glyphosate Herbicide.Plants (Basel). 2023 Jun 12;12(12):2288. doi: 10.3390/plants12122288. Plants (Basel). 2023. PMID: 37375914 Free PMC article.
-
Identification of Potential FDA-Approved Inhibitors of SARS-CoV-2 Helicase Through a Multistep In silico Approach: A Promising Prospect for COVID-19 Treatment.Med Chem. 2025;21(5):425-441. doi: 10.2174/0115734064318640241112071225. Med Chem. 2025. PMID: 40525420
-
Arctigenin from Forsythia viridissima Fruit Inhibits the Replication of Human Coronavirus.Int J Mol Sci. 2024 Jul 4;25(13):7363. doi: 10.3390/ijms25137363. Int J Mol Sci. 2024. PMID: 39000469 Free PMC article.
-
Integrated study of Quercetin as a potent SARS-CoV-2 RdRp inhibitor: Binding interactions, MD simulations, and In vitro assays.PLoS One. 2024 Dec 3;19(12):e0312866. doi: 10.1371/journal.pone.0312866. eCollection 2024. PLoS One. 2024. PMID: 39625895 Free PMC article.
References
-
- WHO WHO Coronavirus (COVID-19) Dashboard. [(accessed on 10 September 2021)]. Available online: https://covid19.who.int/
-
- Xu J., Hagler A. Chemoinformatics and drug discovery. Molecules. 2002;7:566–600. doi: 10.3390/70800566. - DOI
-
- Jalmakhanbetova R.I., Suleimen Y.M., Oyama M., Elkaeed E.B., Eissa I., Suleimen R.N., Metwaly A.M., Ishmuratova M.Y. Isolation and In Silico Anti-COVID-19 Main Protease (Mpro) Activities of Flavonoids and a Sesquiterpene Lactone from Artemisia sublessingiana. J. Chem. 2021;2021:5547013. doi: 10.1155/2021/5547013. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

